tRNA Definition
Transfer RNAs or tRNAs are molecules that act as temporary carriers of amino acids, bringing the appropriate amino acids to the ribosome based on the messenger RNA (mRNA) nucleotide sequence. In this way, they act as the intermediaries between nucleotide and amino acid sequences.
tRNAs are ribonucleic acids and therefore capable of forming hydrogen bonds with mRNA. Additionally, they can also form ester linkages with amino acids, and therefore, can physically bring mRNA and amino acids together during the process of translation. They pair with mRNA in a complementary and antiparallel manner, and each tRNA can base pair with a stretch of three nucleotides on mRNA. These sets of three nucleotides on the mRNA are called codons and the corresponding sequence on the tRNA is called the anticodon. Base pairing between the codon and anticodon brings specificity to the process of translation. On one end of the tRNA, an appropriate amino acid is attached to its 3’ hydroxyl group based on the anticodon and the ribosome catalyzes the formation of a peptide bond between this amino acid and the elongating polypeptide chain.
tRNA Structure and Function
Transfer RNAs are coded by a number of genes, and are usually short molecules, between 70-90 nucleotides (5 nm) in length. The two most important parts of a tRNA are its anticodon and the terminal 3’ hydroxyl group, which can form an ester linkage with an amino acid. However, there are other aspects to a tRNA’s structure such as the D-arm and T-arm, which contribute to its high level of specificity and efficiency. Only 1 in 10,000 amino acids are incorrectly attached to a tRNA, which is a remarkable number given the chemical similarities between many amino acids.
Transfer RNAs have a sugar-phosphate backbone like all other cellular nucleic acids and the orientation of the ribose sugar gives rise to directionality in the molecule. One end of the RNA has a reactive phosphate group attached to the fifth carbon atom of ribose while the other end has a free hydroxyl group on the third carbon atom. This gives rise to the 5’ and 3’ ends of the RNA since all the other phosphate and hydroxyl groups are involved in phosphodiester bonds within the nucleic acid.
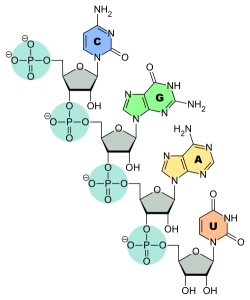
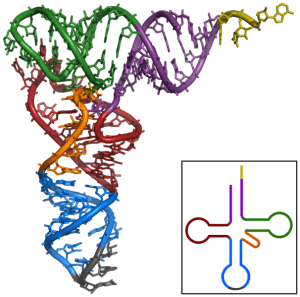
The last three bases on the 3’ end of tRNA are always CCA – two cytosines followed by one adenine base. This stretch is part of the acceptor arm of the molecule, where an amino acid is covalently attached to the hydroxyl group on the ribose sugar of the terminal adenine nucleotide. The acceptor arm also contains parts of the 5’ end of the tRNA, with a stretch of 7-9 nucleotides from opposite ends of the molecule base pairing with each other.
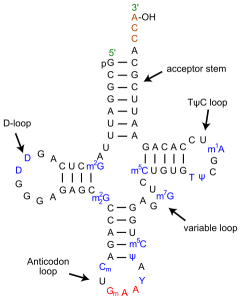
The anticodon loop, which pairs with mRNA, determines which amino acid is attached to the acceptor stem. The anticodon loop is recognized by aminoacyl tRNA synthetase (AATS), the enzyme that chemically links a tRNA to an amino acid through a high-energy bond. AATS ‘reads’ the anticodon and also recognizes the D-arm located downstream from the 5’ end of the tRNA.
The D-arm is made of a double-stranded stem region formed by internal base pairing as well as a loop structure of unpaired nucleotides. The D-arm is a highly variable region and plays an important role in stabilizing the RNA’s tertiary structure and also influences the kinetics and accuracy of translation at the ribosome.
The other structure that influences the role of tRNA in translation is the T-arm. Similar to the D-arm, it contains a stretch of nucleotides that base pair with each other and a loop that is single stranded. The paired region is called the ‘stem’ and mostly contains 5 base pairs. The loop contains modified bases and is also called the TΨC arm, to specify the presence thymidine, pseudouridine and cytidine residues (modified bases). tRNA molecules are unusual in containing a high number of modified bases as well as containing thymidine, usually seen only in DNA. The T-arm is involved in the interaction of tRNA with the ribosome.
Finally, a variable arm containing less than 20 nucleotides is situated between the anticodon loop and the T-arm. It plays a role in AATS recognition of tRNA, but could be absent in some species.
The secondary structure of tRNA containing the acceptor region, D- and T-arms and the anticodon loop is said to resemble a cloverleaf. After the RNA folds into its tertiary structure, it is L-shaped, with the acceptor stem and T-arm forming an extended helix and the anticodon loop and D-arm similarly making another extended helix. These two helices align perpendicularly to each other in a way that brings the D-arm and T-arm into close proximity while the anticodon loop and the acceptor arm are positioned on opposite ends of the molecule.
In this image, the 3’ CCA region is in yellow, the acceptor arm is in purple, the variable loop in orange, the D-arm is in red, the T-arm in green and the anticodon loop is in blue.
Types of tRNA
A tRNA can be classified based on the amino acid it carries, giving rise to 20 different tRNAs. Alternatively, they can also be grouped based on their anticodon. There are 64 possible codons arising from a combination of four nucleotides. Of these, 3 are stop codons that signal the end of translation. This gives rise to a situation where one amino acid is represented by multiple codons and the AATS, as well as the tRNAs have to accommodate this redundancy. However, very few species have exactly 61 tRNAs, which gives rise to the question of how every codon is recognized by a specific tRNA. In many species, the number far exceeds 61 and different tRNAs carrying the same anticodon could display varying efficiency in translation, adding a layer of regulation to the process of protein synthesis.
tRNAs interact with codons on the mRNA through their anticodon loop. Base pairing between the codon and anticodon ensures specificity during translation. However, the first base of the anticodon, that pairs with the ‘wobble’ or third position in a codon is often modified to allow the tRNA to hydrogen bond with three, instead of one base. Thus a single tRNA has the option of recognizing and base pairing with three codons, which code for the same amino acid. There are 20 AATS, one for each amino acid. This group of enzymes can recognize all the anticodons representing a particular amino acid and therefore act as the second arm of the machinery that handles genetic code redundancy.
Finally, these molecules can also be classified into three categories – those carrying canonical amino acids attached to the correct tRNA, those that are incorrectly attached, and those carrying modified amino acids such as selenocysteine for non-canonical elongation.
Post-Transcriptional Modification of tRNA
There are nearly 500 genes coding for tRNAs in the human genome, and 300 gene fragments associated with these RNA. These genes are transcribed by RNA polymerase III and the transcript undergoes extensive modification, especially in eukaryotes. Introns are spliced, the intron-exon boundary is acted on by endonucleases, the 5’ and 3’ ends of the RNA are processed and enzymes add the terminal CCA residues to the 3’ end of the tRNA. The CCA residues could become aminoacylated in the nucleus itself and this charged tRNA could then be exported from the nucleus.
Additionally, many bases on the tRNA are also modified, especially by methylation (addition of a methyl group) and deamidation (removal of an amide group). Particularly, the first base of the anticodon that pairs with the ‘wobble’ position on the codon is modified to allow unusual types of base pairing. Adenine can be modified to form inosine, which expands the pairing possibilities to include uracil, cytosine and adenine. Pseudouridine is another common modified base, derived from uridine residues through enzyme-mediated isomerization. It is said to play a role in the structural integrity of the tRNA molecule, being involved in stiffening the nearby sugar-phosphate backbone and also influencing base stacking of proximal regions. Lysidine is an unusual base formed when a lysine amino acid is attached to cytidine residue. Lysidine pairs specifically with adenosine, a property that is used by the isoleucine tRNA to ensure translation specificity.
AATS attach the appropriate amino acid to tRNA molecules based on their anticodon. These enzymes contain binding sites for the amino acid, tRNA as well as ATP and hydrolyze ATP to AMP and attach the amino acid to the ribose sugar of the last nucleotide on tRNA. The tRNA is now considered ‘charged’ and can participate in the protein synthesizing reactions on the ribosome. This reaction often occurs in the cytoplasm, though it has also been observed in the nucleus.
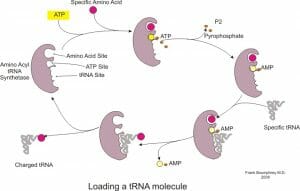
The enzyme binds to many regions of the tRNA to ensure high specificity in the reaction and even proofreads its own reaction since many amino acids have similar structures.
Mature tRNA then binds specific export factors that export it from the nucleus, using the RanGTP system. The acceptor arm and T-arm play an important role in this process, and there is extensive interaction between the export factors and the RNA molecule, allowing only fully processed, complete tRNAs to move to the cytoplasm.
tRNA Interaction with Ribosome
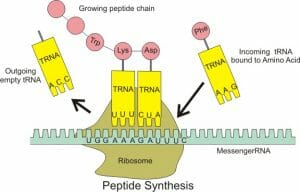
The ribosome contains three important regions – the P (peptidyl) site containing the growing polypeptide, the A (acceptor) site that receives a new charged tRNA and the E (exit) site through which a deacylated tRNA leaves the ribosome. These sites span both the subunits of the ribosome and are denoted as P/P or A/A sites with the first letter referring to the site on the smaller subunit. For instance, the P/P site binds to tRNA anchoring a polypeptide chain while the A/A site anchors an incoming charged tRNA. The peptidyl-tRNA on the P/P site transfers the growing polypeptide to the tRNA on the A/A site and undergoes deacylation. To continue the process of translation, the ribosome moves ahead by one codon, making the tRNA on the P/P site shift to a transient P/E configuration and then to the E/E site before leaving the ribosome. Similarly, the tRNA on the A/A site adopts a temporary A/P binding conformation before settling at the P/P site, ready for the next amino acid to continue translation.
Related Biology Terms
- Antiparallel – Parallel but running in opposite directions, such as the two sugar-phosphate backbones of a DNA molecule.
- Complementarity – The property of nitrogenous bases in nucleic acids to form specific and stable hydrogen bonds with other nitrogen bases. For instance, the interaction between adenine and thymine is through complementary hydrogen bonding.
- Introns – Parts of an RNA molecule that are removed post-transcriptionally.
- Transcription – The process of generating an RNA molecule from a DNA template.
Quiz
1. Which of these is a structure found on tRNAs?
A. Anticodon loop
B. Codons
C. AATS
D. All of the above
2. Which of these modified residues are found on tRNAs?
A. Pseudouridine
B. Thymidine
C. Cytidine
D. All of the above
3. Which of these modified bases is derived from adenine?
A. Adenosine
B. Inosine
C. Cytidine
D. All of the above