Exon Definition
An exon is a coding region of a gene that contains the information required to encode a protein. In eukaryotes, genes are made up of coding exons interspersed with non-coding introns. These introns are then removed to make a functioning messenger RNA (mRNA) that can be translated into a protein.
Exon Structure
Exons are made up of stretches of DNA that will ultimately be translated into amino acids and proteins. In the DNA of eukaryotic organisms, exons can be together in a continuous gene or separated by introns in a discontinuous gene. When the gene is transcribed into pre-mRNA the transcript contains both introns and exons. The pre-mRNA is then processed and the introns are spliced out of the molecule. Mature mRNAs can be a few hundred to several thousand nucleotides long.
The mature mRNA consists of exons and short untranslated regions (UTRs) on either end. The exons make up the final reading frame which consists of nucleotides arranged in triplets. The reading frame begins with a start codon (usually AUG) and ends in a termination codon. The nucleotides are arranged in triplets as each amino acid is coded for by a three-nucleotide sequence.
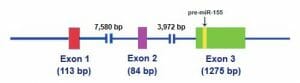
The figure depicts a gene made up of three exons. The resulting gene is 1317 bp in length despite an initial gene length of over 13,000 bp.
Exon Function
Exons are pieces of coding DNA that encode proteins. Different exons code for different domains of a protein. The domains may be encoded by a single exon or multiple exons spliced together. The presence of exons and introns allows for greater molecular evolution through the process of exon shuffling. Exon shuffling occurs when exons on sister chromosomes are exchanged during recombination. This allows for the formation of new genes.
Exons also allow for multiple proteins to be translated from the same gene through alternative splicing. This process allows the exons to be arranged in different combinations when the introns are removed. The different configurations can include the complete removal of an exon, the inclusion of part of an exon, or the inclusion of part of an intron. Alternative splicing can occur in the same location to produce different variants of a gene with a similar role, such as the human slo gene, or it can occur in different cell or tissue types, such as the mouse alpha-amylase gene. Alternative splicing, and defects in alternative splicing, can result in a number of diseases including alcoholism and cancer.
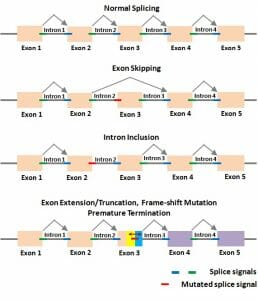
The figure depicts possible alternative splicing mechanisms which can result in alternative proteins being translated.
Human slo gene
An example of extreme alternative splicing is the human slo gene which encodes a transmembrane protein involved in regulation of potassium entry in the hair cells of the inner ear, resulting in frequency perception. The gene consists of 35 exons which can combine to form over 500 mRNAs through the excision of one to eight exons. The different mRNAs control which sound frequencies can be heard.
Mouse alpha-amylase gene
The mouse alpha-amylase gene encodes two different mRNAs – one in the salivary glands and one in the liver. Which of the mRNA transcripts is formed is controlled by different promoters specific to the tissue type. In this case the processed mRNA contains the same two exons, resulting in the same protein, but it is regulated by a tissue-specific promoter.
Quiz
1. A protein is coded for by how many exons?
A. 1
B. 2
C. 10
D. All of the above
2. How can new genes be formed?
A. Alternative splicing
B. Exon shuffling
C. Splicing
D. None of the above
3. What sequence is commonly found at the beginning of an exon?
A. AUG
B. UAG
C. UAA
D. UGA
References
- Brown, T. (2012). Introduction to genetics: a molecular approach Ch. 5. New York, NY: Garland Science, Taylor & Francis Group, LLC. ISBN: 978-0-8153-6509-9.
- Klug, W. S., & Cummings, M. R. (1994). Concepts of genetics, 4th ed. Ch. 17. Englewood Cliffs, NJ: Prentice Hall, Inc. ISBN: 0-02-364801-5.
- Warden, A. S., & Mayfield, R. D. (2017). Gene expression profiling in the human alcoholic brain. Neuropharmacology 122: 161-174.
