CRISPR Definition
Clustered regularly interspaced short palindromic repeats (CRISPR) are short, repeating stretches of DNA found in most archaea and many bacteria. CRISPR and CRISPR-associated proteins (Cas) form an adaptive immune system that protects against foreign genetic elements such as viruses, plasmids, and transposons. The CRISPR/Cas9 system also provides the basis for a genome editing tool that can be used to permanently modify genes in a specific, targeted manner.
Features of CRISPR Loci
CRISPR loci are composed of an AT-rich leader sequence followed by multiple, short nucleotide repeats separated by spacer regions. Many of the repeats are palindromic, with predicted RNA hairpin secondary structure, while others lack symmetry and are predicted to form unstructured RNA. Repeats are conserved within a CRISPR locus, while the spacers are highly variable and correspond to DNA acquired from exposure to foreign DNA, such as viruses and plasmids. The length of CRISPR repeats and spacers varies; repeats are 23-55 basepairs long, and spacers are 21-72 basepairs long. CRISPR arrays are usually located adjacent to clusters of Cas genes. The Cas genes encode a wide variety of proteins that have functional domains found in helicases, nucleases, polymerases and other nucleotide-binding proteins. Cas proteins have functions in acquiring spacer DNA, RNA processing, and target binding and cleavage. CRISPR loci have been identified in approximately 90% of Archaea and 40% of Bacteria, and an organism’s genome may contain one or many CRISPR loci (up to 18 have been observed in a single organism).
CRISPR Classes and Types
There are two classes of CRISPR/Cas systems, termed Class 1 and Class 2. Class 1 systems utilize multiple Cas proteins to cleave foreign DNA, while Class 2 systems use a single Cas protein. The classes are divided into types; Class 1 contains types I, III, and IV, and Class 2 contains types II, V, and VI. Types are defined by a gene generally found only within that type, as well as the additional Cas genes present. The types are further divided in 19 subtypes.
Function and Mechanism of CRISPR in Prokaryotes
CRISPR provides prokaryotes with acquired immunity against invasive genetic elements. These loci incorporate genetic material from viruses and plasmids, and use it to target foreign genetic elements in a sequence-specific manner. The steps involved in protecting against foreign genetic elements using the CRISPR/Cas system are acquisition of spacer DNA from the invading virus, biogenesis of CRISPR RNA (crRNA), which will allow for recognition of foreign DNA, and interference, in which the invasive DNA is recognized and cleaved.
Acquisition of Spacer DNA
When a prokaryotic cell is invaded by a virus, portions of the viral DNA are sampled and incorporated into the spacer regions of the CRISPR locus. The nuclease enzymes Cas1 and Cas2 are involved in spacer acquisition in E. coli and likely all CRISPR/Cas systems, as they are the only Cas proteins found to be conserved across all systems. The DNA that is sampled and incorporated into CRISPR loci as spacers may be located next to protospacer adjacent motifs (PAMs) within the viral genome; PAMs are important in selection of foreign nucleic acids in type I and type II systems. Spacers are usually added next to the leader sequence, though the new spacer may also be inserted randomly into the repeat-spacer array.
crRNA Biogenesis
Following incorporation of foreign DNA into a CRISPR locus, the CRISPR sequence is transcribed and processed into small interfering crRNAs. The repeat-spacer array is initially transcribed into a single transcript, which is then processed by Cas proteins into crRNAs, which later serve as a guide in targeting foreign nucleic acids. Biogenesis of crRNA differs in different CRISPR systems. In certain type I systems, Cas proteins cleave at the edge of stem-loops in the long transcript to produce smaller crRNAs. In type II systems, a trans-activating crRNA (tracrRNA) is transcribed that is complementary to the CRISPR repeats, resulting in the formation of double stranded RNA (dsRNA). The dsRNA is cleaved by RNaseIII to produce crRNAs.
Interference
During the interference stage, crRNAs associate with Cas proteins to form a complex that recognizes, targets, and destroys viral genetic material. The crRNA basepairs with the complementary sequence in the viral DNA, marking it for destruction. Recognition of the PAM sequence may also be required for recognition of the foreign DNA. In type II systems, Cas9 carries out the interference step using both a crRNA and tracrRNA which allows it to recognize foreign DNA. In addition to recognizing target sequences, Cas9 has endonuclease activity and cleaves the foreign DNA.
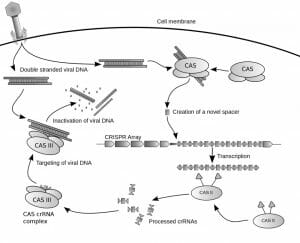
The figure depicts how the CRISPR/Cas system defends against foreign genetic elements in prokaryotes.
CRISPR/Cas9 System as a Genome Editing Tool
The CRISPR/Cas system has been exploited as a molecular biology technique for targeted genome editing. Genome editing is carried out using CRISPR/Cas9 system. The Cas9 protein is a DNA endonuclease that uses a guide RNA to target and cleave DNA. Native Cas9 utilizes a guide RNA composed of crRNA and trans-activating CRISPR RNA (tracrRNA); for the purposes of genome editing, this system was simplified by fusing crRNA and tracrRNA to form a single guide RNA. The CRISPR/Cas9 system may also include a repair template, which is utilized in DNA repair via non-homologous end joining or homology directed repair. By altering the sequence of the guide RNA, the CRISPR/Cas9 system can be used to target any DNA sequence, and knockdown, activate, or mutate the desired sequence. The components of the CRISPR/Cas9 system are often incorporated into a plasmid that is used to transfect cells for genome editing. Multiple changes can be made simultaneously; in one case 62 genes were modified at once.
Gene Activation and Repression
CRISPR that lacks nuclease activity (termed dCas9) can be used to target a specific sequence for activation or suppression. By eliminating the ability to cut DNA, the CRISPR/Cas9 system can be used to target genes without cleaving them. Cas9 lacking nuclease activity alone blocks transcription in bacterial cells; in mammalian cells additional proteins are incorporated. Transcription factors may also be coupled to Cas9, allowing for activation of target genes.
Altering the Genetic Sequence
A DNA repair template can be included in the CRISPR/Cas9 system which allows for this DNA sequence to be incorporated at the desired location. The repair template extends beyond the location of the cleaved section of DNA. Once the DNA break is introduced, the cell’s DNA repair machinery uses the template to repair the DNA, thereby permanently altering the sequence of the DNA.
CRISPR Applications
CRISPR technology can be used to make precise genetic alterations in diverse cell types and organisms, including mice, plants, fish, and human cells. CRISPR can be used to generate animal models of disease by knocking down or targeting a gene of interest. CRISPR can also be used to generate cell lines that contain disease-causing mutations and can be use to study the molecular mechanisms of the disease. CRISPRs also have promising therapeutics applications; they may be effective in targeting viruses such as herpesvirus and preventing their replication in humans, have been used to treat genetic disorders in animals, and have been approved for clinical trials in the treatment of cancer. Apart from biomedical applications, CRISPR can also be used to engineer biofuel-producing yeast strains as well as food crops.
Quiz
1. The CRISPR/Cas system provides adaptive immunity against foreign genetic elements in what types of organisms?
A. Bacteria
B. Archaea
C. Eukaryotes
D. A and B
2. What does Cas9 do after binding DNA that is complementary to its guide RNA?
A. Replicates it
B. Cuts it
C. Transcribes it
D. All of the above
3. Which of the following is not a feature of CRISPR?
A. Contain palindromic DNA repeats
B. Have highly conserved spacer regions
C. Provide adaptive immunity to Bacteria and Archaea
D. Used as a molecular biology tool for genome editing
References
- Chen, K.Y. and Knoepfler, P.S. (2016). “To CRISPR and beyond: the evolution of genome editing in stem cells.” Regenerative Medicine. 11(8):801-816.
- Doetschman, T. and Georgieva, T. (2017). “Gene Editing With CRISPR/Cas9 RNA-Directed Nuclease.” Circulation Research. 120(5):876-894.
- Patterson, A.G., Yevstigneyeva, M.S., and Fineran, P.C. (2017). “Regulation of CRISPR-Cas adaptive immune systems”. Current Opinion in Microbiology. 37:1-7.
- Salsman, J. and Dellaire, G. (2017). “Precision genome editing in the CRISPR era.” Biochemistry and Cell Biology. 95(2):187-201.
