Transcription Definition
Transcription refers to the first step of gene expression where an RNA polymer is created from a DNA template. This reaction is catalyzed by enzymes called RNA polymerases and the RNA polymer is antiparallel and complementary to the DNA template. The stretch of DNA that codes for an RNA transcript is called a transcription unit and could contain more than one gene.
These RNA transcripts can either be used as messengers to drive the synthesis of proteins or be involved in a number of different cellular processes. These functional or non-coding RNA could be transfer RNA (tRNA), ribosomal RNA (rRNA), or direct gene regulation through RNA interference (RNAi) and the formation of heterochromatin.
Function of Transcription
Life on earth is said to have begun from self-replicating RNA since it is the only class of molecules capable of both catalysis and carrying genetic information. With evolution, proteins took over catalysis because they are capable of a greater variety of sequences and structures. Additionally, the bonds on the sugar phosphate backbone of RNA are vulnerable to even mild changes in pH and can undergo alkaline hydrolysis. Therefore, DNA emerged as the preferred molecule for carrying genetic information since it is more stable and resistant to degradation. Transcription maintains the link between these two molecules and allows cells to use a stable nucleic acid as the genetic material while retaining most of their protein synthesis machinery.
In addition, separating DNA from the site for protein synthesis also protects genetic material from the biochemical and biophysical stresses of complex, multilayered processes. Small errors in the RNA transcript can be overcome since the RNA molecule has a short half-life, but changes to the DNA become heritable mutations. In addition, transcription adds another layer for intricate gene regulation, especially in species with large genomes that require minute adjustments in metabolism.
In eukaryotes, transcription also plays an important role in transferring the information from DNA to the cytoplasm because the nuclear pore is too small to allow ribosomes, proteins or chromosomes to pass through. While the nuclear pore has a diameter of about 5-10 nm, ribosomes are between 25-30 nm in size, many proteins are wider than 10 nm and fully condensed chromosomes can be over 2000 nm in size. Therefore, the primary machinery for protein synthesis cannot enter the nucleus and stretches of DNA cannot exit the nucleus.
Mechanism of Transcription
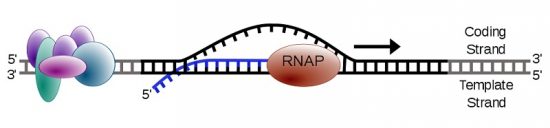
Transcription creates a single stranded RNA molecule from double stranded DNA. Therefore, only the information in one of the strands is transferred into the nucleotide sequence of RNA. One strand of DNA is called the coding strand and the other is the template strand. Transcription machinery interacts with the template strand to produce an mRNA whose sequence resembles the coding strand. Other names for the template strand include antisense strand and master strand.
Two different genes on the same DNA molecule can have coding sequences on different strands.
Transcriptional activity is particularly high in the G1 and G2 phases of the cell cycle when the cell is either recovering from mitosis or preparing for the dramatic events of the next cycle of cell division.
Transcription Initiation
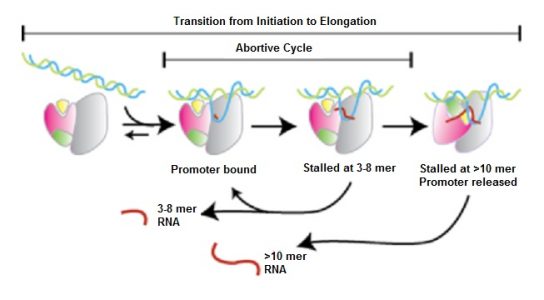
Transcription begins with the binding of an RNAP in the presence of general transcription factors to the promoter region upstream of the transcription start site on the DNA. Prokaryotic RNAP binds with a sigma factor, while eukaryotic RNA polymerases can interact with a number of transcription factors as well as activator and repressor proteins. Initially, after the binding of RNAP to the promoter region, the DNA remains in a double-stranded form. This is called a ‘closed complex’ between DNA and RNAP. Thereafter, RNAP along with transcription factors unwinds a segment of the DNA and interacts with the exposed nucleotides in an open complex creating a ‘transcription bubble’. RNAP then cruises along the DNA scanning for the transcription start site inside the bubble. Once the start site is located, the first two nucleotides of the transcript are bonded to each other.
Escape from Promoter
After the first few nucleotides are added to the putative RNA transcript, RNAP enters a critical, unstable phase. It can either continue towards productive initiation, or pull DNA towards itself, creating scrunched open DNA inside the polymerase. If RNAP rewinds the downstream portion of the DNA, the putative RNA transcript is released because the DNA-RNAP complex reverts to its initial open configuration. This is called abortive initiation.
However, if the upstream portion of DNA is rewound and ejected from the enzyme, RNAP moves ahead. Its interaction with the promoter region is broken and the RNA transcript reaches a length of 14-15 nucleotides. This is called escape from the promoter and is accompanied by changes to protein-protein and protein-DNA interactions. Some transcription factors are released and transcription moves towards the elongation phase.
Transcription Elongation
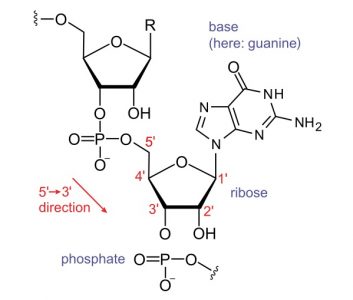
Once a short RNA oligonucleotide of more than 15 bases is formed, RNAP proceeds along the template DNA strand. The transcript is identical to the coding strand, except that the nucleotide backbone has ribose sugar instead of deoxyribose, and adenine base pairs with uracil, instead of thymine. RNAP can catalyze the formation of a phosphodiester bond between the fifth carbon atom of an incoming nucleotide and the third carbon atom of the last nucleotide in the existing transcript.
Since the RNA molecule has a free phosphate attached to the fifth carbon on the first nucleotide and a free hydroxyl group on the third carbon of the last nucleotide, RNA is said to be transcribed in a 5′ to 3′ direction.
Transcription Termination
Unlike DNA replication, where the DNA polymerase continues to add nucleotides till it reaches the end of the molecule, transcription has to be terminated at a particular location for effective gene regulation and expression. Prokaryotic transcription termination can occur through the formation of a double-stranded region within the RNA or through the action of a protein called Rho.
The first method involves the transcription of a G:C rich region followed by a string of uracils that form weak hydrogen bonds with template DNA. The G:C rich region can loop over itself to form a hairpin-like structure stalling the RNAP and transcription machinery. This, combined with the weaker bonds between uracil and the template DNA can prise the RNA away from the transcription machinery and lead to termination. This process also involves a protein called NusA.
Rho-dependent transcription termination involves the binding of Rho protein to a sequence on the transcribed RNA. This sequence, which is downstream from translation stop codons, allows Rho to bind to RNA and cruise along the transcript in an ATP-dependent manner. When it encounters a stalled RNAP, it binds to the enzyme and causes the transcript and its associated machinery to dissociate from the DNA.
Eukaryotic transcription termination is much less understood, and most of the work has focussed on the mechanisms of RNAP II. Transcription termination in eukaryotes is also coupled with post-transcriptional modifications and processing before the mature RNA is exported to the cytoplasm.
Types of RNA Transcripts
Traditionally, three types of RNA transcripts were known – messenger RNA (mRNA), tRNA and rRNA – and all three are intimately associated with protein synthesis . While mRNA determines amino acid sequence, tRNA and rRNA are crucial for the mechanism of translating the mRNA code.
mRNA polymerization from DNA containing protein coding genes is catalyzed by RNA polymerase II. Occasionally, proteins that are used together are coded as a single unit, in one long mRNA molecule and this is particularly common among prokaryotes. DNA sequences upstream of the coding sequence contain docking sites for the transcription machinery as well as regulatory factors that modulate the timing and quantity of transcriptional activity. mRNA is then modified and processed to give rise to the final transcript used for translation.
rRNA constitutes nearly fifty percent of the RNA of a cell and is transcribed by RNA polymerase I in specialized regions of the nucleus called the nucleolus. Nucleoli appear as dense spherical structures around the loci that code for rRNA. Prokaryotic rRNA is of three types and eukaryotic ribosomes are made of four types of rRNA with the largest one containing over 5000 nucleotides. These RNA molecules determine the three-dimensional structure of ribosomes.
RNA polymerase III catalyzes the transcription of tRNA precursors in the nucleus. Promoter sequences controlling the expression of tRNA genes can be intragenic, located inside the coding sequence of the gene. tRNA precursors undergo extensive modifications including splicing. Prokaryotic tRNAs retain their catalytic activity and can self-splice, whereas eukaryotic post-transcriptional modification is carried out by special endonuclease enzymes. These endonucleases recognize specific structural motifs within the tRNA that target the sequence for splicing.
In addition to these three types of RNA, the cell contains a number of smaller RNA involved in various cellular activities. These include gene regulation (mediated by micro RNA and sequences in the 5′ untranslated regions of mRNA transcripts), post-transcriptional modification (small nuclear RNA, small nucleolar RNA), genome defense (Piwi-interacting RNA and CRISPR) and the maintenance of genomic structure (telomeres and RNA transcripts that silence X-chromosomes).
Differences between Prokaryotic and Eukaryotic Transcription
The obvious difference between prokaryotic and eukaryotic transcription is the presence of a nuclear membrane in eukaryotes. Eukaryotic RNA transcripts need to be exported from the nucleus, whereas prokaryotes conduct coupled transcription and translation in the cytoplasm. This is possible because the prokaryotic transcript does not undergo extensive modification and prokaryotes do not need transcription factors for initiation. Therefore, the transcription machinery is simpler and can simultaneously accommodate the enzymes of translation.
Prokaryotes also have only one RNA polymerase to catalyze all the transcription reactions of the cell and a single RNA transcript can direct the synthesis of multiple proteins. These mRNA are called polycistronic mRNA. Often, all the genes involved in one biochemical pathway are transcribed and translated together, allowing the entire pathway to be regulated as a single unit. In eukaryotes, polycistronic mRNA can be found in chloroplasts.
Related Biology Terms
- Monocistronic mRNA – mRNA transcript that codes for a single protein.
- Transposons – Small segments of DNA that can move around the genome, inserting themselves into loci far removed from their original site, often involving an RNA intermediate.
- hnRNA – Heterogenous nuclear RNA are considered the original products of transcription and consist mostly of mRNA precursors.
- Poly-A polymerase – Enzyme that adds a stretch of adenine nucleotides to the end of a primary transcript.
Quiz
1. Which of these properties makes DNA a more stable genetic material?
A. The hydrogen bonds between the bases are stronger
B. DNA is longer than RNA
C. Presence of thymine bases
D. Resistance to degradation through alkaline hydrolysis
2. What is the size of a nuclear pore in eukaryotes?
A. Less than 10 nm
B. More than 10 nm
C. Over 2000 nm
D. 25-30 nm
3. Which of these is NOT a feature of prokaryotic gene expression?
A. Coupled transcription and translation
B. Extensive post-transcriptional modification of the RNA transcript
C. Sigma factor for transcription initiation
D. None of the above