Definition
DNA polymerase is an important enzyme group involved in DNA synthesis, repair, and replication; these enzymes are found in all living organisms. Originally discovered during research into Escherichia coli bacteria, we now know of multiple varieties with similar structures but different functions. These varieties are grouped into families according to function and are also used in the field of genetic engineering.
DNA Polymerase Function
DNA polymerase has varying roles in the mechanisms of DNA synthesis, repair, and replication. DNA polymerase is categorized into seven different families in eukaryotes, viruses, yeasts, and bacteria. These seven families are A, B, C, D, X, Y, and reverse transcriptase (RT). Future research may discover further groups.
Each of these families contains a subset of DNA polymerases that have their own range of functions. For example, DNA polymerase I is a member of the A family; DNA polymerase IV or DinB is a member of the X family. You will not need to memorize every name, but the basic function per group will also help you to better understand protein synthesis, gene mutation, and gene modification.
The structure of DNA polymerase is likened to a right hand with a palm, fingers, and thumb. You can imagine a strand of DNA moving through a DNA polymerase molecule like the ribbon through a typewriter. Very simply put, the fingers help to carefully position the unzipped DNA strand by recognizing the nucleotides, the palm is the active site where phosphorylation occurs (adding the phosphate backbone), and the thumb binds the DNA into a double-helix form as it exits the DNA polymerase molecule. But not all DNA polymerase families have the same structural components. Let’s look at the different families in a little more detail.
Polymerase Family A
Family A is a group of DNA replication or DNA repair enzymes. In DNA replication, they match a nucleotide base to the right partner. This is necessary whenever a cell prepares to divide, and the single-stranded chromosome is duplicated so that both cells, mother and daughter, have a full set of DNA.
If a copy of the DNA is to be made, DNA polymerase molecules run over the unzipped template strand and copy it with opposite nucleotides. This produces an exact copy of the coding strand of DNA. Different family A enzymes help toward DNA repair – they check newly-produced strands for faulty bases and replace them if faults are found.
Examples of family A DNA polymerases are Pol I, Pol γ (gamma), and Pol θ (theta). Often referred to as the Pol I family (Pol is short for polymerase), each sub-type has a specific action.
You can always tell whether a DNA polymerase is found in prokaryotic or eukaryotic cells by looking at their names. When a polymerase is allocated Roman numerals (Pol III, Pol I, and so on), that enzyme is found in prokaryote (single-celled) organisms. In eukaryotes, sub-types are named according to the Greek alphabet (Pol delta, Pol theta, and so on). Families can contain DNA polymerases for single and/or multi-celled organisms.
Pol γ is the only DNA polymerase that can replicate mitochondrial DNA (and only family X DNA polymerases carry out mtDNA repair).
Pol theta (DNA polymerase theta) repairs double-strand breaks within the DNA by rejoining the broken ends. Damage to the gene that codes for Pol theta (θ) production means that breaks begin to pile up without getting repaired; however, theta-mediated end joining (TMEJ) increases the risk of mutation when compared to some other DNA repair mechanisms. Because of this, faulty Pol θ genes have been linked to many forms of cancer.
Because of such studies into disease and DNA, family A DNA polymerases have helped us to understand and treat various forms of cancer. Another A-family example is Pol nu which helps to unhook interstrand cross-links (ICL). What is an interstrand cross-link? Have you ever heard of mustard gas, used in World War II? Breathing this gas in large quantities could kill, but thousands of soldiers survived exposure. As time went by, doctors found these brave men were more likely to die of respiratory system cancers than people who had never been exposed to mustard gas. The gas entered the lungs and reacted directly with the DNA of lung cells, tying together one nucleotide strand to opposite nucleotides that were not their partners (diagonal- or cross-links). These extra bonds made unzipping DNA before replication difficult and, when replication did occur, mistakes were made in copying the code. These mistakes multiplied over time, causing many DNA faults that were copied and caused gene mutations. These mutations led to the production of faulty cells or cancer. In the case of mustard gas, this was lung cancer.
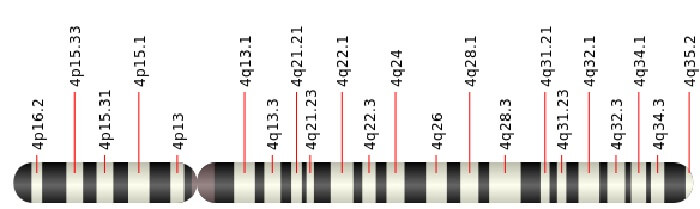
Pol nu (POLν) is specifically produced to try to solve these highly-damaging interstrand crosslinks. It is not made in great quantities and seems to be more of a back-up enzyme, but there may be more to it than that. Although only discovered in 2003, lesser-known DNA polymerases like Pol nu are receiving a lot of attention. One of the reasons is that around 50% of breast cancer cells show deleted areas at cytogenic location (position) 4p16.2 – that’s chromosome 4, short arm (p), region 16, band 2). In the below image, it is the position furthest to the left. Also important to note is that this is exactly where the gene for Pol nu synthesis is located.
Polymerase Family B Function
DNA polymerase B family enzymes are important during the process of cell division. They check newly-replicated and synthesized DNA. The family includes both prokaryote and eukaryote polymerases.
Pol alpha (a Greek letter, so a eukaryote polymerase) kick-starts the DNA replication process and communicates areas of damage to other B-family DNA polymerases such as Pol delta and Pol epsilon. Because these faults are immediately fixed, they are much more likely to be successful and the risk of mismatch repair (matching the wrong nucleotide to a damaged strand of DNA) is low.
An example of mismatch repair is the replacing of a previously bonded guanine and thymine pair to produce a guanine and cytosine pair in the DNA, where thymine is wrongly substituted with cytosine. Bacterial and eukaryote DNA polymerases are central to both damage recognition and damage repair mechanisms.
Polymerase Family C Function
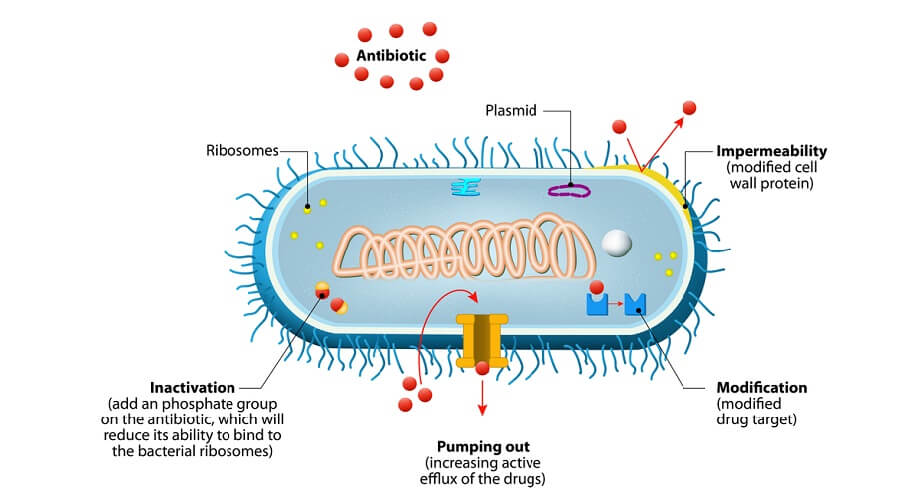
While DNA polymerase C functions are only found in bacteria, we should never forget that bacteria outnumber human cells by ten to one on and within the average body. The majority of these are essential for our health, assisting the digestive system, and producing chemicals that improve system and organ function. Less often, pathogenic bacteria colonize to produce symptoms of illness and disease. Family C – often referred to as PolC – is the most important bacterial DNA replication polymerase group. Family C is not a repair polymerase.
With drug-resistant bacteria on the rise, new antibacterial agents are becoming more and more necessary. New areas of research include developing antibiotics that directly target PolC. This potential new broad-spectrum drug could prevent replication in all types of bacteria, healthy and pathogenic, but even more importantly, these drugs – that are still in the earliest stages of development – avoid the mechanisms that lead to bacterial antibiotic resistance.
Polymerase Family D Function
Euryarchaeota describes a group of gram-positive and gram-negative bacteria that are often said to prefer extreme environments (extremophiles). However, these bacteria live and multiply in all kinds of environments, from deep marine silts to our digestive systems. They use D-family DNA polymerases (PolD) for DNA replication. Mutation rates in this group are very high when compared to those of PolB DNA polymerases. And unlike other polymerases, family D does not have a hand-like structure, probably because these cells are, evolutionarily speaking – very early cell types.
Polymerase Family X Function
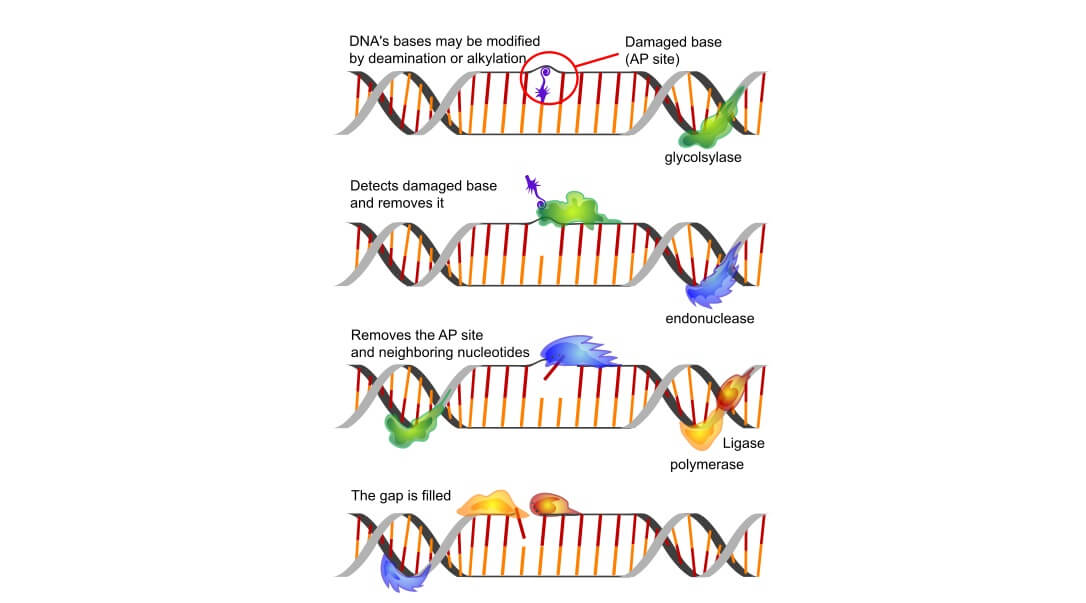
The X family of DNA polymerase is limited to eukaryotic cells and plays both replicative and repair roles. Some work to repair mitochondrial DNA where high oxidative environments encourage DNA damage. Others repair one to (approximately) ten consecutive nucleotides in the DNA of the cell nucleus. The method of repair (base excision repair) in the mitochondrion and nucleus is similar. Base excision repair (BER) is a process that uses various types of enzymes, including DNA glycosylase and endonucleases. It is X-family DNA polymerase (Pol beta and Pol lambda) that forms the active site for this repair and inserts the correct nucleotide. If the gene for X-family DNA polymerases is damaged, BER processes are negatively affected and this is associated with certain types of cancer. Some new targeted therapies developed for these cancers inhibit faulty base excision repair mechanisms.
Polymerase Family Y Function
The DNA polymerase Y family is a replicative and repair enzyme found in eukaryotic and prokaryotic cells. All of these polymerases are very error-prone in respect to their role in the replication and immediate repair or bypassing of faulty DNA sequences. Yet at the same time, too low levels of this family of polymerases can increase one’s susceptibility to malignant tumors. This is why the Y family is sometimes likened to a double-edged sword.
The Y-family group activates when other DNA polymerases are unable to make an effect. It is supposed to be a backup mechanism; this may explain why mutations following this type of repair are more common.
Reverse Transcriptase Function
Viruses, retroviruses, and eukaryote cells contain RNA-dependent reverse transcriptase enzymes. These enzymes – part of the DNA polymerase group – are what make viruses dangerous. As a virus only contains RNA, it must trick a microorganism or cell into reproducing it. If our cells only copied the RNA, they might produce one or two unusual proteins in a ribosome but these would not help the virus to multiply. Instead, the viral RNA must somehow make itself part of the DNA template so that the cell undergoes permanent changes. It does this by using reverse transcriptase enzymes.
These enzymes produce double-stranded DNA from a single-stranded RNA template in a process known as reverse transcription. Mutations are common. The image below shows how the human immunodeficiency virus replicates in a T-lymphocyte. Reverse transcription initiates growth of the virus by tricking the cell into producing components that assemble to form more viruses from edited DNA.
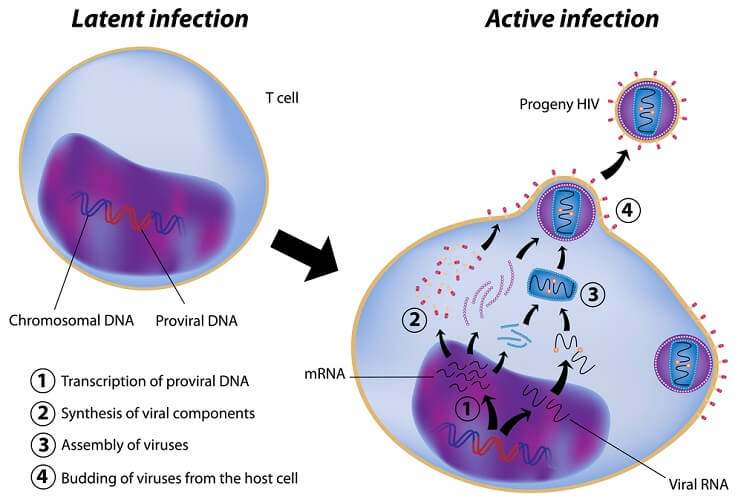
Most reverse transcription processes are the result of harmful viral infections, where the single-stranded viral RNA is copied to form a double DNA strand that will go on to make viral proteins. This is done by reverse transcriptase (reverse because the usual method is to use double-stranded DNA to produce a single strand of RNA).
Testing during the COVID-19 (SARS-CoV-2) in 2020 – as with all viral infection testing – requires viral RNA extraction. Laboratories use a process called the reverse transcriptase-polymerase chain reaction (rt-PCR). Rt-PCR isn’t as complicated to understand as it might sound. This test produces complementary DNA (cDNA) or DNA that is copied from small amounts of viral RNA. As this procedure only produces very small quantities of cDNA, the results must be amplified by replicating it. Once produced in sufficient quantities, the viral genome can be detected.
