Translation Definition
Translation refers to the process of creating proteins from an mRNA template. The sequence of nucleotides on the RNA is translated into the amino acid sequence of proteins and this reaction is carried out by ribosomes. Ribosomes and tRNA dock on a mature mRNA transcript and recruit multiple enzymes in an energy intensive process that uses ATP as well as GTP.
Genetic Code
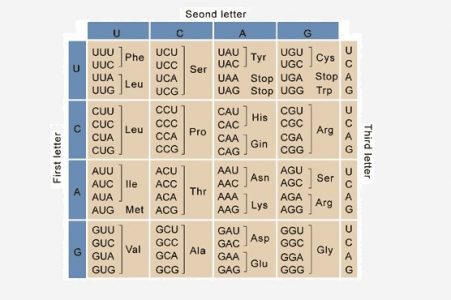
When nucleic acids were discovered as the primary genetic material, one important question came to the fore. There are only 4 bases in nucleic acids whereas proteins are made of 20 amino acids. Therefore, it is not possible to have a direct one-to-one correlation between the sequences of nucleotides and amino acids. Even having two nucleotides code for a single amino acid is insufficient. Therefore, it was suggested that amino acids were coded by stretches of three nucleotides called codons. A series of experiments in the 1960s confirmed this hypothesis and also showed that these codons do not overlap with each other. In addition, since stretches of 3 nucleotides can give rise to a total of 64 codons, a single amino acid can be coded by multiple codons, a property that is called ‘degeneracy’. Often, the difference between degenerate codons is the third base which is called the ‘wobble position’. For instance, the amino acid serine can be coded by six codons of which four are: UCA, UCG, UCU or UCC. Similarly, phenylalanine can be represented by either UUU or UUC on the mRNA and leucine is coded by a total of six codons. This degeneracy is aided by the fact that the third nucleotide in every codon binds loosely to its corresponding tRNA, allowing unusual types of bases to pair with each other.
Out of the 64 codons made by various combinations of 4 nucleotides, 3 are stop codons, that signal the end of translation. These are UAA, UAG and UGA and are recognized by proteins called release factors rather than by tRNAs. When a ribosome encounters a stop codon, it dissociates from the mRNA through the enzymatic action of release factors.
mRNA Untranslated Regions
The entire stretch of mature mRNA does not consist of codons that are translated into amino acid sequence of a protein. There is a ‘cap’ to the 5′ end of the RNA, two short stretches of untranslated regulatory regions abutting the coding sequence (the 5′ UTR and 3′ UTR), and a polyadenylate tail that can determine protein sequence without directly being translated.
These regions are involved in the export of mRNA from the nucleus, protection from enzymatic degradation, and regulation of translational activity. They contain binding sites for proteins that can enhance or reduce translation, docking sites for ribosomes and other parts of the translation machinery, as well as enzymes that catalyze the degradation of mRNA when the protein requirement of the cell is met.
tRNA Structure and Function
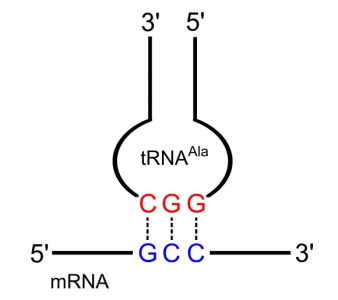
Transfer RNA act as adapter molecules between mRNA and amino acids, bringing the appropriate amino acid to the ribosome based on mRNA codons. tRNAs contain a three-base anticodon that can recognize and bind with mRNA as well as act as a signal for the correct amino acid. The anticodon sequence is complementary to the mRNA codon, and runs in an antiparallel direction, allowing the two molecules to base pair with each other.
A group of enzymes called aminoacyl tRNA synthetases attach the appropriate amino acid to tRNA molecules based on their anticodon. There is one aminoacyl tRNA synthetase for each of the 20 amino acids and the enzyme can recognize all the anticodons that represent that particular amino acid. These enzymes use the energy from ATP to attach the amino acid to the last nucleotide on the 3′ end of tRNA. The tRNA is now considered to be ‘charged’ and can participate in the protein synthesizing reactions on the ribosome.
Ribosome Structure and Function
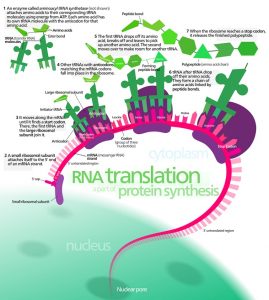
Ribosomes are macromolecular, multi-subunit structures containing RNA as well as protein and are the primary machines that drive protein synthesis. The structure of the ribosome derives primarily from it RNA component (ribosomal RNA or rRNA) and base-pairing with mRNA and tRNA is crucial to its function.
The ribosome contains two subunits and translation is initiated when the smaller subunit binds to sequences upstream of the coding sequence on the mRNA. Prokaryotic translation begins with the rRNA directly binding to the mRNA, whereas eukaryotic translation involves other proteins called initiation factors. The smaller subunit, along with some other proteins recruit the larger subunit of the ribosome, and translation begins.
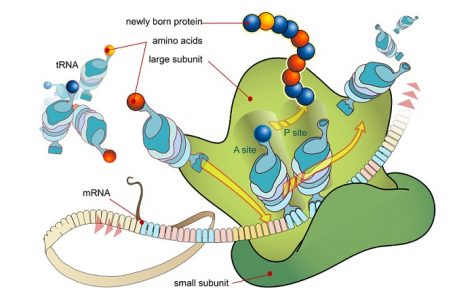
Primarily, the ribosome contains three important regions – the P site, the A site and the E site – formed by the three-dimensional shape of the rRNA. The P site binds to the growing polypeptide, the A site anchors an incoming charged tRNA and after peptide bond formation, the tRNA binds briefly to the E site before leaving the ribosome.
Translation Mechanism
Translation proceeds in three stages – initiation, elongation and termination. Each of these is associated with different proteins and at every step, ATP and GTP are used as energy sources.
A single mRNA can be translated by multiple ribosomes in a process called translatome. These complexes were initially called ergosomes and are now called polysomes or polyribosomes.
Initiation
Translation begins with the mature mRNA transcript being exported from the nucleus and its 5′ cap being recognized by the smaller subunit of a ribosome. The ribosomal subunit, along with a special tRNA, scans the mRNA to find the start site for translation, which is often AUG – the codon for methionine. The sequences around the AUG start codon are also important and can determine how strongly an mRNA is translated. Initiation also involves the activity of a number of helper proteins called initiation factors, whose function is to ensure that the various parts of the translation machinery come together in an orderly manner.
The ribosome and initiator tRNA move slowly along the mRNA till the start codon is located. The structural features of this tRNA ensure that it is recognized by initiation factors and discriminated against by translation elongation factors. The methionine attached to this tRNA is also adapted to be exclusive for initiation – the amino group is modified to make N-formyl methionine, preventing it from participating in the elongation phase of translation.
Once the larger subunit of the ribosome arrives at the translation start site, and the tRNA is located at its P site, initiation is said to be complete.
Elongation
Binding of the initiator tRNA to the P site on the ribosome brings it into close contact with the amino or ‘A’ site of the ribosome, where the next codon awaits translation. New tRNAs carrying amino acids enter the ribosome at the A site. Through complementary base pairing and the energy from one molecule of GTP, the correct tRNA binds to the ribosome. Ribosomal RNA then catalyzes the formation of a peptide bond between the first and second amino acids where the first methionine appears to have been ‘transferred’ to the tRNA on the A site. Once the peptide bond is formed, the empty tRNA exits the ribosome and the ribosome itself moves ahead by exactly one codon, so that the tRNA on the A site moves into the P site. This also exposes the next codon to the A site, ready for the third tRNA. This process continues till a stop codon is reached.
Termination
When a stop codon is present at the A site it is recognized by a set of proteins called release factors. They induce the ribosome to attach a molecule of water to the growing polypeptide chain, rather than another amino acid. This terminates the process of translation, and releases the polypeptide from the ribosome. The two subunits of the ribosome also dissociate from each other, ready for the next cycle of translation.
Translation on the Endoplasmic Reticulum
Translation can occur either on free ribosomes in the cytoplasm or on ribosomes present on the surface of the endoplasmic reticulum (ER). Translation begins in the cytoplasm for nearly all proteins. However proteins that are needed as internal membrane proteins or those that need to be secreted from the cell are targeted for further translation on the ER. These proteins contain a short stretch of hydrophobic residues called the signal peptide at the start of their sequence. As soon as these residues are translated, the signal peptide is recognized by specific proteins called signal-recognition particles, which can transport the entire ribosome and associated molecules to the membrane of the ER. The signal peptide gets embedded in the ER membrane and the rest of the protein is released into the interior space of the ER. The signal peptide is removed from proteins that need to be secreted from the cell, while those intended for internal membranes retain that short stretch providing a membrane anchor.
Occasionally, proteins needed inside organelles like mitochondria and chloroplasts are translated in the cytoplasm. These proteins are selectively transported into these organelles using specific proteins in an energy intensive process.
Antibiotic Targets
The differences between prokaryotic and eukaryotic translation machinery make them ideal drug targets for combating infections, while leaving the body’s cells intact. These antibiotics include chloramphenicol, tetracycline, puromycin and erythromycin. However, since most animals also contain a rich inner biome of symbiotic bacteria, these antibiotics can also cause a number of side effects, including vitamin deficiencies.
Related Biology Terms
- 3′ -> 5′ orientation – Directionality of a single strand of nucleic acid which derives from the numbering of carbon atoms on the nucleotide sugar ring. One end of the nucleic acid has a free hydroxyl group on the third carbon and the other end has a free phosphate group attached to the fifth carbon.
- Antiparallel orientation – Two biopolymers that run parallel to each other but in opposing orientations. The two strands of a DNA are antiparallel to each other, with respect to the orientation of their sugar-phosphate backbones.
- Energy currency of the cell – Small nucleotides containing high energy bonds that are used for storing and releasing energy within the cell. ATP and GTP are common energy currencies in the cell.
- Rough Endoplasmic Reticulum – Endoplasmic Reticulum that is studded with ribosomes, where proteins that are intended for internal membranes or for secretion are translated.
Quiz
1. Which of these codons codes for serine?
A. UCA
B. UAA
C. UAG
D. All of the above
2. Which of these enzymes is involved in translation?
A. Aminoacyl tRNA synthetase
B. 23S rRNA
C. Peptidyl Transferase
D. All of the above
3. How many aminoacyl tRNA synthetases exist in the cell?
A. 64 – one for each codon
B. 61 – one for each codon excluding stop codons
C. 20 – one for each amino acid
D. 1 – a single enzyme catalyzes all the reactions