Definition
Restriction enzymes, restriction endonucleases, or molecular scissors are bacteria-produced enzymes that can slice between two DNA strands at areas called recognition sites. Restriction enzymes were first discovered during Enterobacteria coli research. Type II restriction enzymes (REs) are of particular importance in the fields of molecular cloning, gene sequencing, and DNA mapping as this group can cut DNA very close to specific recognition sites and does not require energy in the form of ATP.
Restriction Enzyme Function
Restriction enzyme function in the natural world is to defend bacteria against specific viruses called bacteriophages. These viruses attack bacteria by injecting viral RNA or DNA into a bacterial plasmid (small, purple ring in the below image) and replicating there. If viral RNA or DNA is detected within a prokaryote cell, that cell can often stop the replication process by slicing through the foreign genetic information. This renders it useless. At the same time, bacterial DNA is protected from the cutting action of its restriction endonucleases within its restriction sites. This mechanism adds methyl (H3C) groups to the cytosine and adenine of bacterial DNA without affecting the coded DNA sequence.
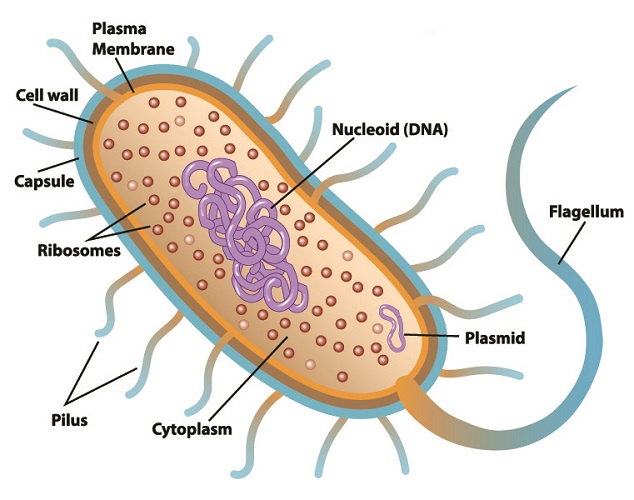
To date, approximately 3500 restriction enzymes have been isolated from bacterial plasmids. Each enzyme recognizes a specific sequence of viral genetic code and will try to separate the new, mutated DNA strand close to or further away from the recognition site. This natural separation mechanism is also referred to as restriction enzyme digestion.
Not only the location and the method but also the type of cut can differ. Some REs leave uneven sticky ends (non-blunt ends) between slightly different areas of a double-strand that overhang; others leave blunt ends where base pairs are separated at the same point. For example, BamHI is a type II restriction enzyme obtained from Escherichia coli that recognizes the nucleotide sequence GGATCC and cleaves these sections of DNA leaving sticky ends. Cleaving, like cleaving a log with an ax, is the scientifically-accepted term for cutting a strand of DNA.
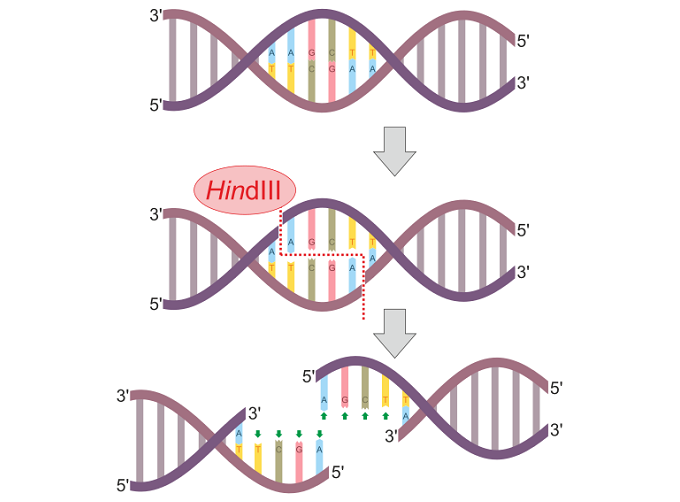
In the below image, a restriction enzyme called HindIII cleaves DNA at different points on the two strands to form a sticky end.
Once the double DNA strand has been separated, another enzyme called DNA ligase rejoins the DNA backbone as a sticky-end or blunt-end ligation. Single-stranded DNA that has been inserted into bacterial DNA by specific viruses can be removed by certain REs. DNA ligase then recombines the DNA by constructing a mirror copy of the bacterial sequence. In short, a restriction enzyme cleaves the foreign DNA and DNA ligase repairs the break to bring it back to its original form.
Since the discovery of genes, ways to manipulate them have been heavily researched. The action of removing a gene sequence and replacing it with another is known as gene recombination. The short restriction enzyme recognition sites usually number between four to eight nucleotides. This makes restriction enzymes ideal for use in the field of molecular biology.
Nucleotides in DNA consist of a nucleobase, a deoxyribose sugar, and a phosphate group. It is the phosphate and sugar groups that form the backbone of DNA, shown here in blue and turquoise.

Restriction Enzyme Groups
Natural restriction enzymes are arranged in five groups: type I, II, III, IV, and V. Type I REs, the first to be discovered, cut DNA sequences far from the recognition sites and require ATP to recognize, modify and/or digest asymmetrical sections. The distance from the recognition site makes type I restriction endonucleases less helpful in the field of genetic engineering.
Type II REs recognize and cut short sections of DNA close to restriction sites without ATP but using magnesium ions. Because of this, they are the most commonly used natural restriction endonucleases. Type II restriction enzymes are further categorized into subgroups and one of these subgroups is the high-precision IIS category.
Type III restriction endonucleases are rarely used in genetic engineering as they cut DNA sequences well outside of the recognition sequence and need to detect two separate sequences to achieve this. This means they are not always able to provide complete restriction enzyme digestion. Type IV restriction nucleases can only cleave methylated DNA (DNA that is not transcribed into a protein) and sequence specificity is weak.
Type V restriction enzymes require guide RNA (gRNA) to target specific sequences and it is these that are being modified or used in genome engineering methods such as TALENS and CRISPR-Cas9. CRISPR is the short form of clusters of regularly interspaced short palindromic repeats. CRISPR regions refer to repeated nucleotide and spacer patterns within a section of the DNA; it is within spacers that viruses incorporate their DNA. By inserting other genetic code into a spacer by artificial means it is possible to modify the genome of a living organism.
Finally, artificial restriction enzymes (AREs) are becoming ever more popular with geneticists as they can be modified to recognize and cut DNA sequences at predefined sites. CRISPR and TALENS use adapted restriction enzymes for increased accuracy; they can also edit many genes in a single process. Furthermore, commercially-available natural restriction enzymes are limited in number, and these fragment DNA into very short sections; it is rare that a smaller laboratory has access to the right enzymes. This is because different restriction enzymes are required to cleave the many separate areas of DNA that make up the code for a single gene.
The recent synthesis of artificial restriction enzymes using certain proteins such as Argonaute protein (PfAgo) provides an alternative technique that can cleave longer sticky-end sequence sequences with increased accuracy.
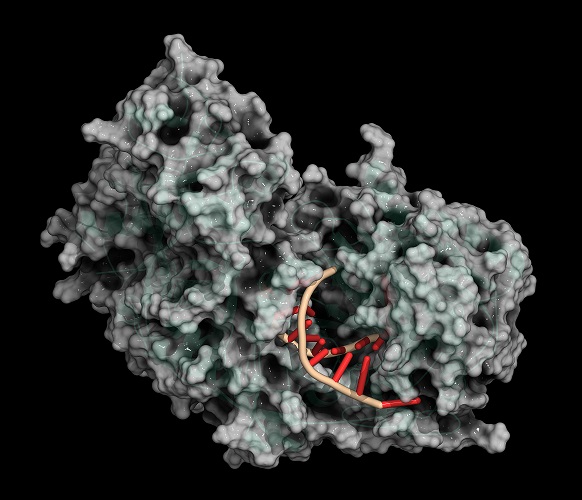
Restriction Enzymes in Molecular Cloning
In molecular cloning, molecular biologists insert a gene into a small, stable section of an organism’s DNA, allowing it to be replicated. When this gene is expressed, research on that gene’s effects on study organisms can be carried out.
Restriction enzyme cloning is one of the earliest techniques in the field of molecular cloning but remains popular due to a low cost-to-reliability ratio. After producing sticky or blunt ends, cleaved DNA is purified and inserted into the DNA of the host bacteria in a step called transformation. After transformation, the plasmid contains recombinant (recombined) DNA – a term used to describe the combination of extracted DNA fragments with DNA ligase enzymes. DNA ligase allows this section to be fixed into a plasmid. A host bacterium can then produce more DNA or express the inserted gene after protein synthesis.
Newer methods that do not require natural restriction enzymes but use synthetic versions are being increasingly implemented. The above-described technique is, therefore, commonly referred to as traditional cloning. DNA cloning should not be confused with the process used to create Dolly the sheep; only small strands of DNA are replicated in gene modification. Dolly was the result of complete genome cloning.

Restriction Enzymes in DNA Profiling
The discovery of restriction enzymes has made DNA profiling possible. Minisatellites are short, repetitive sequences of between ten and sixty base pairs that show greater variation between individuals than other sequences within the genome. This variation is determined by the number of repeated units (stutters) within a minisatellite sequence.
Multiple minisatellites provide a DNA fingerprint that identifies an individual. Earlier forms of DNA profiling used natural restriction enzymes to cut various-sized sections throughout the DNA. Pulsed-field gel electrophoresis separates these sections ready for identification. The separated sections representing minisatellites are blotted onto a membrane and pulled apart to produce single strands. This procedure requires opposing strands composed of radioactive phosphorous that link to their complementary (matching) strands on the membrane. An x-ray then produced an image of the DNA fingerprint – an image is possible due to the radioactive phosphorus copy.

Today, microsatellites of two to five base pairs are replicated many times over through a technique known as the polymerase chain reaction. This newer method provides results even with a tiny sample of DNA – something the earlier method was unable to do. Instead of radioactive phosphorous, primer RNA binds to both ends of those cut DNA sequences that show the most variation between individuals. RNA primers are labeled with fluorescent colors. Pulsed-field gel electrophoresis, multilocus sequence typing (MLST), and polymorphic amplified typing sequences (PATS) are technologies used to separate the resulting fragments. Lasers then provide different light wavelengths to produce a colorful DNA fingerprint.
While DNA profiling is most associated with the field of criminal forensic science, this identification method is also used to detect bacterial strains responsible for disease, provide a bacterial fingerprint that can be used to isolate and treat infection, or determine whether food or places where food is produced is free of pathogenic bacteria.
Traditional DNA Cloning Uses
Traditional DNA cloning using restrictive endonucleases has multiple uses. Expressed recombinant DNA (DNA sequences that code for protein synthesis), when inserted into the genetic information of bacteria, stimulate bacteria to produce the target protein.
This is the method whereby genetic engineers in pharmaceutical companies manufacture human insulin, human albumin, some vaccines, monoclonal antibodies, and human growth hormone at much lower cost that extracting these products from multicellular organisms. Recombinant DNA is also used to diagnose hereditary disease and produce antibiotics on a huge scale.
Gene analysis is a broad sector in which genetic engineers insert cleaved recombinant DNA sequences (rDNA) to help us understand what specific genes do. By understanding genes, we then have the data we need to make adjustments that can potentially eradicate disease.
Genetically-modified crops are the result of traditional molecular cloning techniques where resistance to insects and herbicides and more product per square hectare are the main goals. This method also improves nitrogen fixation in plants and crops.
The catering industry uses recombinant DNA in fermentation and cheese-making processes, and also to detect the presence of pathogenic bacteria and fungi on surfaces used for food preparation.

However, to produce results that may improve our health or food sources, our knowledge of the function of every gene is essential. Only once the function of a DNA sequence has been discovered can it be correctly used. Traditional DNA cloning was the first technique used in the field of genome mapping that has, over many years, taught us how genes are expressed. With new artificial restriction enzymes, genetic engineering can only be expected to move forward over the next few decades.
