Frameshift Mutation Definition
Frameshift mutations are insertions or deletions in the genome that are not in multiples of three nucleotides. They are a subset of insertion-deletion (indel) mutations that are specifically found in the coding sequence of polypeptides. Here the number of nucleotides that are added or removed from the coding sequence are not multiples of three. They can arise from extremely simple mutations such as the addition or removal of a single nucleotide.
Frameshift mutations do not include substitutions where a nucleotide replaces another. In substitution mutations, the polypeptide only changes by a single amino acid. Frameshift mutations also do not include indels in the non-coding or regulatory regions of the genome because these mutations do not have any direct effect on amino acid sequence, though protein regulation may change.
Effects of Frameshift Mutations
Frameshift mutations are among the most deleterious changes to the coding sequence of a protein. They are extremely likely to lead to large-scale changes to polypeptide length and chemical composition, resulting in a non-functional protein that often disrupts the biochemical processes of a cell. Frameshift mutations can lead to a premature end to translation of the mRNA as well as the formation of an extended polypeptide.
The amino acid sequences downstream of the frameshift mutation are also likely to be chemically distinct from the original sequence. For instance, if a frameshift mutation occurs in an integral transmembrane protein, it could vastly alter the stretch of hydrophobic residues that span the lipid bilayer making it impossible for the protein to be present in its subcellular location. When such errors occur, the cell often perceives the lack of functional protein and tries to compensate by upregulating the expression of the mutated gene. This can even overwhelm the translation machinery of the cell, result in a large number of misfolded proteins that could eventually lead to large-scale impairment of all functions of even cell death.
Diseases caused by frameshift mutations in genes include Crohn’s disease, cystic fibrosis, and some forms of cancer. On the other hand, when some proteins become dysfunctional, they could have a protective effect, as seen in the resistance to HIV in people with a chemokine receptor gene (CCR5) containing a frameshift mutation.
Since frameshift mutations are usually changes to the genetic material in every cell, it is rare to find a cure. Most interventions are palliative.
The Genetic Code
The core reason for the presence of frameshift mutations is the body’s mechanism for translating genetic information into amino acid sequences through a triplet-based genetic code. This means that every set of three nucleotides on an mRNA represents either an amino acid or an instruction to cease translation.
Discovery of the Genetic Code
Mendel’s initial experiments on the transmission of genetic traits pointed towards a discrete physical and chemical entity that carried genetic information. Based on the bulk biochemical analysis of cells, four major components were detected – carbohydrates, fats, proteins and nucleic acids. Any of these components could represent genetic material.
Initial investigations into the chemical nature of the genome hypothesized that proteins, with 20 amino acids, were most likely to carry Mendel’s factors or genes. However, later experiments indicated that nucleic acids were the carriers of genetic information. This presented an interesting difficulty. While nucleic acids had been analyzed chemically as being polymers made of 4 different nucleotides, it wasn’t clear how the information for the dazzling variety of forms and functions in the body could arise from just 4 nucleotides.
Triplet Codon
A little later, the central dogma of molecular biology indicated that most organisms used RNA as the intermediate between DNA and proteins. This brought up the next question of how four bases could carry the information to encode 20 amino acids. If every nucleotide coded for a single amino acid, then only four amino acids could be reliably and reproducibly coded. If every two nucleotides encoded an amino acid, it would still lead to only 16 amino acids. Therefore, a minimum of three nucleotides was needed to code for 20 amino acids.
There are 64 permutations possible from nucleotide triplets where each position in the triplet can be one of 4 nucleotides. These nucleotide triplets were named codons. This also gave rise to the idea of redundancy – every amino acid could be represented by more than one codon triplet. Some experiments also revealed that codons were ‘read’ by the translation machinery as discrete chunks of 3 bases. That is, ribosomes ‘see’ these codons like a series of three-letter words. For instance, if an RNA molecule has the sequence AAAGGCAAG, then it can code for a maximum of 3 amino acids from the 3 codons AAG, GGC, and AAG.
Ribosome Translocation
The ribosome moves forward by three bases after each amino acid has been attached to the growing polypeptide chain. The way the ribosome moves is an important reason why frameshift mutations are deleterious and have disproportionate effects on protein function. For instance, if the ribosome only moved by a single base each time, the previous mRNA containing 9 nucleotides can be read as AAA, AAG, AGG, GGC, GCA, CAA and AAG, giving rise to a polypeptide with 7 amino acids. If ribosome translocation only moved one base at a time, the insertion of a single nucleotide would only result in a small change to the amino acid sequence, and possibly no change at all to polynucleotide length.
Reading Frames
In the previous example, the polynucleotide chain can code for a maximum of 3 amino acids. However, depending on the upstream regions, the stretch cans also result in only 2 amino acids. That is, if the ribosome aligns with AAG or AGG instead of AAA initially, the nucleotide polymer is read in a different manner. This way, depending on the position of the translation start site, any coding sequence can be read in 3 different ways. Since most DNA is made of complementary double strands, it leads to a total of 6 different ‘reading frames’, only one of which results in the correct amino acid sequence for the final protein.
However, when there is an indel mutation, there is a shift in the reading frame downstream of the mutation. This results in a frameshift mutation.
Examples of Frameshift Mutation
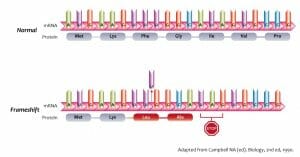
The image above shows the nucleotide and amino acid sequences in a wild type protein as well as the result of a nucleotide insertion, leading to the incorporation of incorrect amino acids and the premature end to polypeptide synthesis. While the original mRNA has a sequence of AUG AAG UUU GGC AUA GUG CCG, the insertion of an extra uracil residue at the ninth position changes the reading frame. Instead of producing a polypeptide of 7 amino acids beginning with methionine and continuing up to proline, it ends after 4 amino acids, with misincorporated leucine and alanine residues.
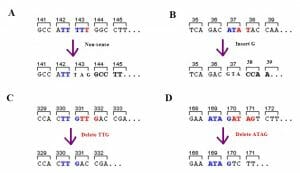
The image below shows the different types of mutations that could severely affect amino acid sequence. Panel A shows the substitution of 2 bases resulting in a premature stop codon, truncating the protein. Panels B and D demonstrate the effect of either the insertion of a single nucleotide or the deletion of 4 nucleotides. In both cases, a frameshift mutation alters all downstream amino acid sequences. Panel C is a subset of indels where 3 (or multiples of 3) nucleotides are inserted or deleted. There is no frameshift mutation. In this particular type of indel mutations, the number of nucleotides mutated is fairly low, there may be very limited effect on protein function as well
Related Biology Terms
- A-site of Ribosome – The ribosomal site that mostly receives an incoming tRNA charged with an amino acid residue. Peptide bonds are formed at the A-site.
- Radiolabeling – Also known as radioisotope labeling, is a technique used to detect the movement of a particular molecule through a chemical, biochemical or cellular system, by replacing some of the atoms in reactants with radioactive isotopes.
- Stop Codons – Nucleotide sequences, especially in mRNA that signal the end of translation. UAA, UAG, and UGA are the canonical stop codons.
- Wild Type – Commonly found strain, gene or characteristic, perceived to have been the original form of the phenotype.
Quiz
1. Which of these would result in a frameshift mutation?
A. Insertion of 3 nucleotides
B. Deletion of 18 nucleotides
C. Insertion of 17 nucleotides
D. All of the above
2. How can a frameshift mutation caused by a single nucleotide drastically change the length of a polypeptide?
A. A change in reading frame changes the position of the translation stop site
B. The insertion or deletion of a nucleotide affects amino acid length
C. The ribosome A-site is unable to proceed beyond the mutation site
D. All of the above
3. Why are frameshift mutations relatively rare?
A. In critical proteins, frameshift mutations can result in non-viable pregnancies
B. They are especially quickly repaired by the DNA repair mechanisms of the cell
C. It is difficult to insert or delete a nucleotide into a DNA stretch
D. All of the above