Definition
DNA replication is a process that occurs during cellular division where two identical molecules of DNA are created from a single molecule of DNA. As a semiconservative process, a single molecule containing two strands of DNA in double helix formation is separated, where each strand serves as a template for the new DNA molecules. Because the double helix is anti-parallel and DNA polymerase only synthesizes new DNA from 5′-3′, the template strand reading 3′-5′ results in a continuous, leading strand, while the template strand reading 5′-3′ results in a discontinuous, lagging strand. Being a highly regulated process, multiple proteins are required both during and following replication to quickly correct mistakes and damages.
Background
Deoxyribonucleic acid, known simply as DNA, is the blueprint of all living things. DNA contains genes that code for the physical and metabolic information expressed in an individual while having the potential to be passed down to future offspring. Almost all cells have DNA, which is typically stored in the nucleus. Notable cells that lack DNA include anucleate cells (or cells that lack a nucleus, such as red blood cells). Additionally, some cells may still have DNA despite not having a nucleus, such as with bacterial cells.
DNA Nucleotides
DNA is made up of four building block monomers that are known as nucleotides. Nucleotides consists of three groups:
- A deoxyribose sugar group
- A phosphate group
- A nitrogenous base
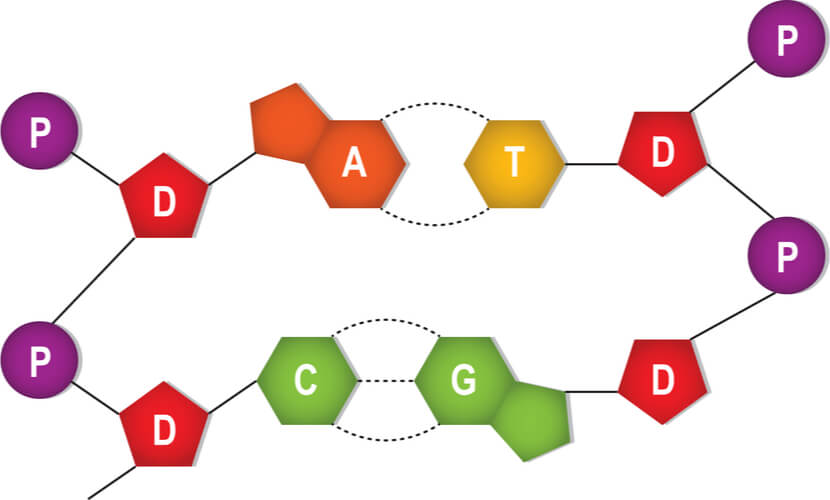
All of the nucleotides have the same sugar group and phosphate group, but different nitrogenous bases. The nitrogenous bases in DNA are adenine, guanine, cytosine, and thymine. Adenine and guanine are classified as purines, while cytosine and thymine are classified as pyrimidines. Purines have two rings in their base structures, while pyrimidines have a single ring in their base structures.
(Helpful hint: A simple pneumonic to remember adenine and guanine as purines is “Pure As Gold”!)
DNA Structure
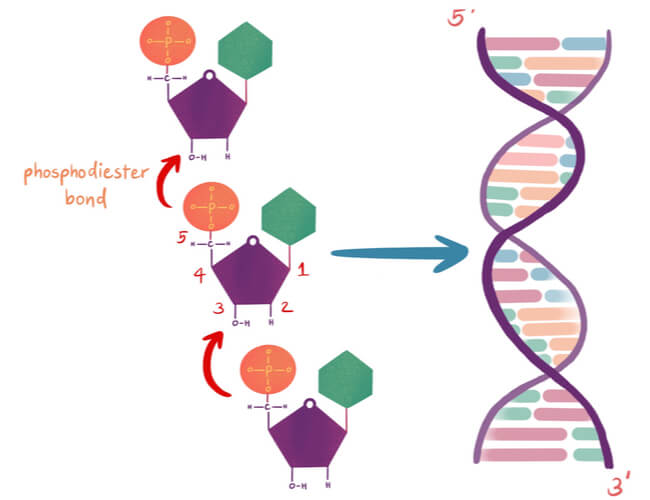
Nucleotides are arranged into chains that become individual strands of DNA, which is half of a full DNA molecule. Each strand has a sugar-phosphate backbone that is created when the phosphate of one nucleotide binds to the sugar of the next using a covalent phosphodiester bond. Specifically, the phosphate is found on the 5′ carbon of one nucleotide, while a hydroxyl group (-OH) is found on the 3′ carbon of the next nucleotide’s sugar group. The -OH of the 3′ carbon is removed, where the phosphate group on the 5′ carbon now also bonds to the 3′ carbon.
The nitrogenous bases stick out from this backbone. A second DNA strand is matched to this first strand based on complimentary base pairing, where a single purine pairs with a single pyrimidine. Specifically, adenine pairs with thymine and guanine pairs with cytosine. Hydrogen bonds connect the complimentary base pairs, where an adenine- thymine pair has two hydrogen bonds and a guanine-cytosine pair has three hydrogen bonds. A single DNA molecule results in double helix formation when two DNA strands are matched and bonded.
DNA has directionality that can run either 3′-5′ or 5′-3′ based off of the carbons in the sugar group. The two strands of DNA in the double helix must run opposite to each other in an anti-parallel fashion. Therefore, if the first strand starts at the 3′ end and finishes at the 5′ end, then the second strand must run opposite, starting at the 5′ end and finishing at the 3′ end.
Example:
3′- AATCGTAA -5′
5′- TTAGCATT -3′
DNA Replication Is Semiconservative
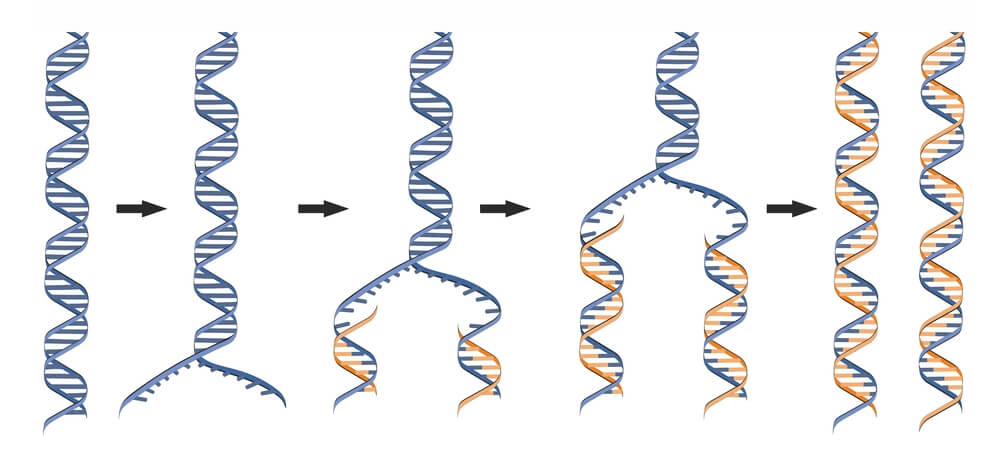
DNA must be fully replicated before cells divide via mitosis to ensure all daughter cells have identical DNA. It was discovered that DNA replication is semiconservative. In semiconservative replication, the double helix splits into two separate strands. During the replication process, an entirely new strand of DNA is created by using the original template strand and matching the complimentary bases.
DNA Replication Process
Proteins in DNA Replication
DNA replication is highly regulated and requires multiple proteins to run efficiently. A majority of these proteins act as stabilizers and enzymes, with enzymes being proteins that behave as catalysts to create and speed up biochemical reactions.
Some of the major proteins in DNA replication include the following:
Helicase: An enzyme that opens the double helix by breaking the hydrogen bonds between complimentary base pairs
Single-strand DNA- binding proteins (SSBPs): These proteins stabilize the individual strands of DNA to prevent them from reconnecting.
Topoisomerase: Because unwinding of the DNA by helicase creates tension further down the strand, this enzyme relieves tension by making cuts in the DNA and rejoining them before the replication fork arrives.
Primase: An enzyme that adds a primer (which is a short segment of ribonucleic acid, known as RNA) where DNA polymerase III will attach
DNA polymerase III: An enzyme that creates the new DNA strand by adding nucleotides that are complimentary to the template strand
DNA polymerase I: An enzyme that replaces the RNA primer with DNA
DNA ligase: An enzyme that connects the Okazaki fragments on the lagging strand by closing the sugar-phosphate backbone, creating a single DNA strand
Sliding clamp: A protein that holds DNA polymerase III in place
The Replication Bubble
When DNA begins to replicate, a replication bubble is formed that can be detected visually by electron microscopy. A specific sequence of bases- known as the origin of replication– determines where this replication bubble begins. Inside of the bubble, two Y-shaped replication forks result where DNA is actively replicated on either side of the region. The replication forks are formed as the double strands of DNA are separated by helicase in both directions away from the origin of replication. It is at the replication fork that DNA replication proteins attach to fulfill their functions.
Replicating the Leading Strand
As mentioned previously, DNA strands have an anti-parallel nature, where one strand will run 3’-5’ and the other will run opposite from 5’- 3’. DNA polymerase can only synthesize new strands of DNA in the 5’-3’ direction. In order for DNA polymerase to do this, it must read the template strand from 3′-5′. Therefore, replicating the template strand that runs 3’-5’ results in the synthesis of the leading strand. The leading strand is a new strand of DNA that is synthesized in a single, continuous chain that starts at the 5’ end and finishes at the 3’ end.
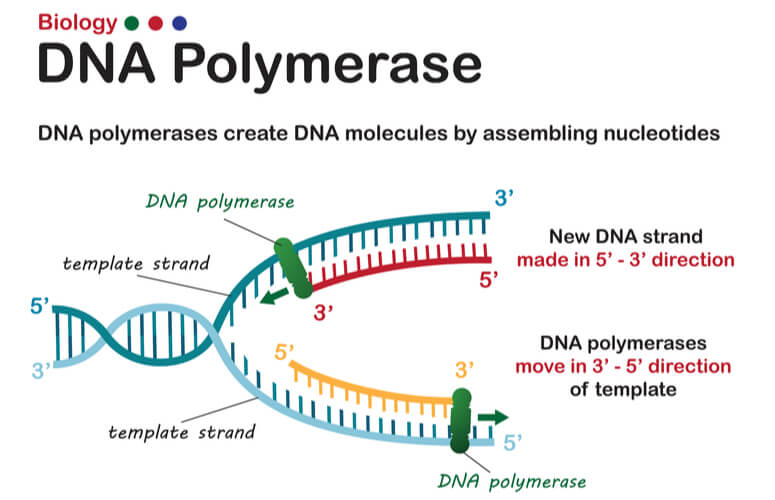
DNA replication of the leading strand when the 3’-5’ template strand is used is as follows:
- The DNA double helix is opened by helicase into individual strands. Topoisomerase relieves the tension further down the double helix.
- SSBPs stabilize the single DNA strands to prevent them from reconnecting.
- An RNA primer is added to the leading strand at complimentary bases by primase.
- DNA polymerase III attaches to the primer. The sliding clamp stabilizes DNA polymerase III.
- DNA polymerase III moves down the leading strand towards the replication fork, adding bases to the new strand from the 5’ end to the 3’ end.
Replicating the Lagging Strand
DNA polymerase can only create new DNA strands from 5’-3’. Therefore, when the 5′-3′ template strand is being replicated- where the new strand must run opposite in the 3′-5′ direction- the new strand cannot be synthesized in a continuous fashion as the leading strand was. To overcome this challenge, additional steps are needed to replicate the 5′-3′ template strand, where this newly synthesized strand is known as the lagging strand.
In order for the lagging strand to be synthesized, DNA needs to be broken down into smaller segments known as Okazaki fragments. Because of these multiple segments, the lagging strand is also known as the discontinuous strand. By creating these multiple segments, DNA polymerase III is able to synthesize a small portion of the new DNA strand away from the replication fork in the correct 5′-3′ direction. As the replication fork continues down the double helix in the 3′ direction of the template strand, another Okazaki fragment can be created closer to the fork. DNA polymerase III binds again to synthesize another portion of the new DNA strand away from the fork until it reaches the previous portion already synthesized. These fragments are then connected, resulting in a single DNA strand. This process continues down the entire length of the DNA.
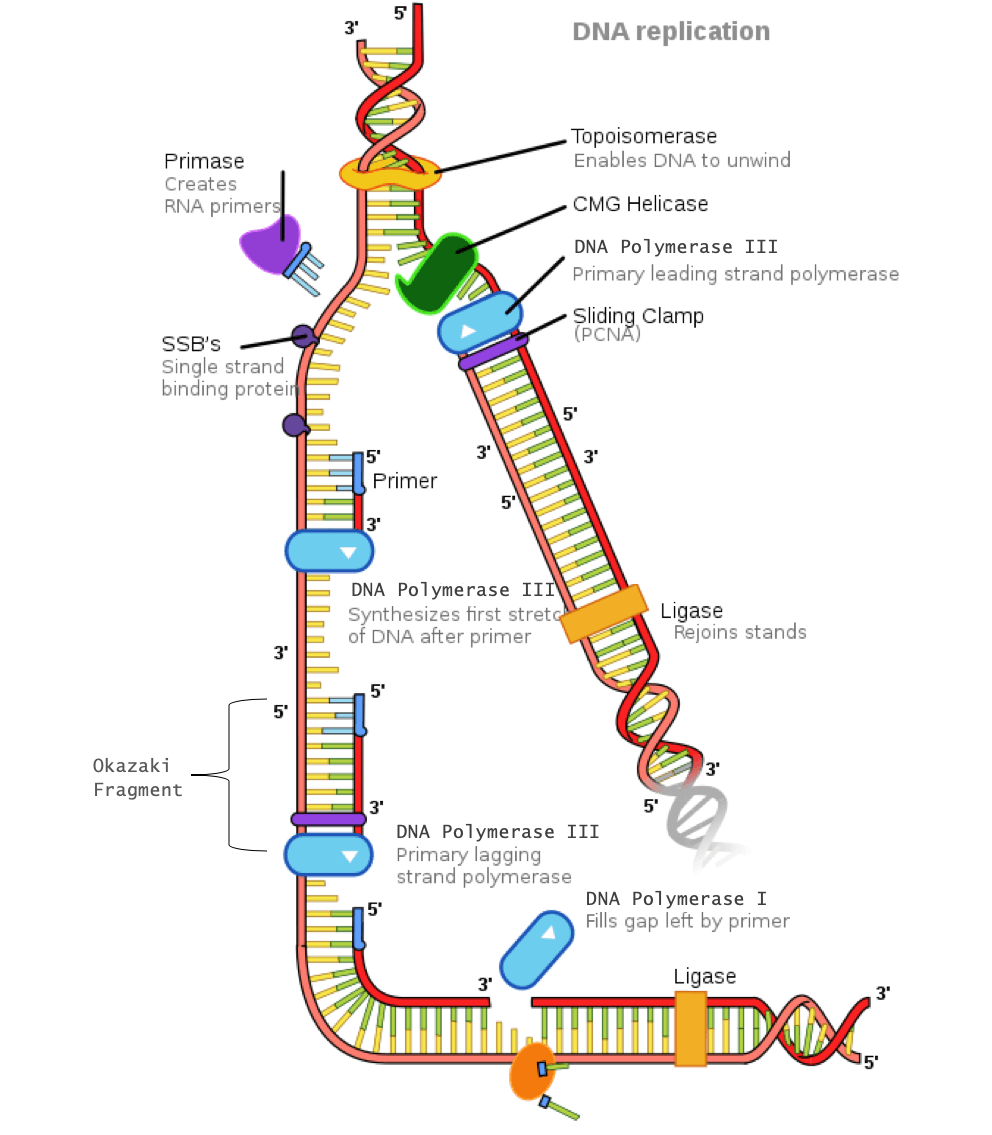
DNA replication of the lagging strand when the 5’-3’ template strand is used is as follows:
- The DNA double helix is opened by helicase into individual strands. Topoisomerase relieves the tension further down the double helix.
- SSBPs stabilize the single DNA strands.
- Primase adds an RNA primer to the lagging strand.
- DNA polymerase III binds to the primer and creates a short segment of newly synthesized DNA from 5′-3′, synthesizing in the opposite direction of the replication fork. This is the first Okazaki fragment.
- As helicase further unwinds the double helix and the replication fork moves down the strand, another primer is added closer to the fork. DNA polymerase III attaches to this primer to synthesize a second Okazaki fragment in the 5′-3′ direction away from the replication fork.
- Once DNA polymerase III reaches the first Okazaki fragment primer, DNA polymerase I removes the primer and replaces them with the proper complementary bases.
- DNA ligase connects the segments of DNA by closing the sugar-phosphate backbone. The two segments are now connected into a single strand.
- This process repeats as the replication fork continues down the length of the DNA.
Prokaryotes vs. Eukaryotes
DNA replication overall is fairly conserved across life. However, general differences exist in the enzymes and mechanisms used, as well as time required between species. The largest differences are between the domains of prokaryotes (bacteria and archaea) and eukaryotes (all other plant and animal cells).
Minor differences between these groups include faster replication time in prokaryotes and shorter Okazaki fragments in eukaryotes. Additionally, prokaryotes only have a single origin of replication, while eukaryotes have multiple origins of replication. The most noteworthy difference between these groups however, is that prokaryotes have circular DNA while eukaryotes have linear DNA. Linear eukaryotic DNA creates an additional challenge that must be regulated. This brings us to telomeres.
Telomeres
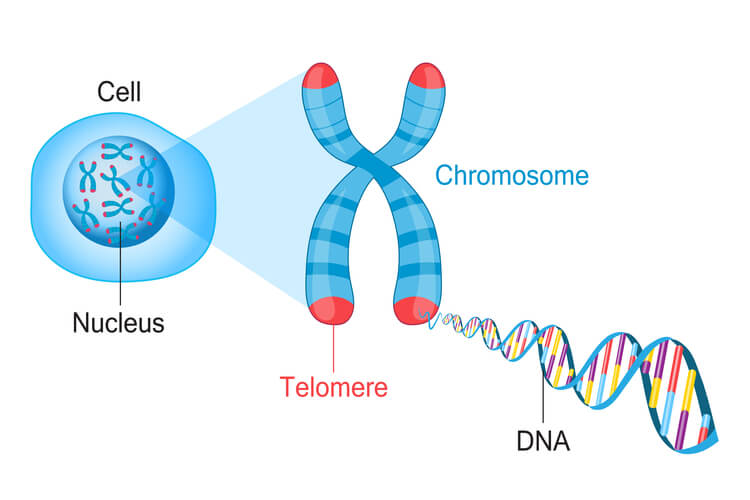
Because eukaryotic DNA is linear, they have ends that create a challenge. For the leading strand, DNA polymerase III can continue down the entire length of DNA. However, in the lagging strand, a primer must be added in front of the Okazaki fragment being synthesized before DNA polymerase III can attach and synthesize the new DNA strand opposite of the replication fork. Once the last Okazaki fragment is synthesized, a small DNA segment is leftover at the tip of the strand. This segment cannot be left unattended. If this DNA isn’t replicated, then genetic material will be lost each time replication occurs. After several replication cycles, this can result in lost information that could be critical for the individual to survive.
To solve this issue, telomeres are present in eukaryotes. Telomeres are short, repeating segments of DNA that are found at the end of each chromosome and do not contain any coding sequences. These telomeres are synthesized by telomerase, which is an enzyme that contains a short RNA template used to extend the length of the lagging strand. Primers are placed on the telomere where DNA polymerase III can attach to synthesize the final portion of DNA leftover on the lagging strand.
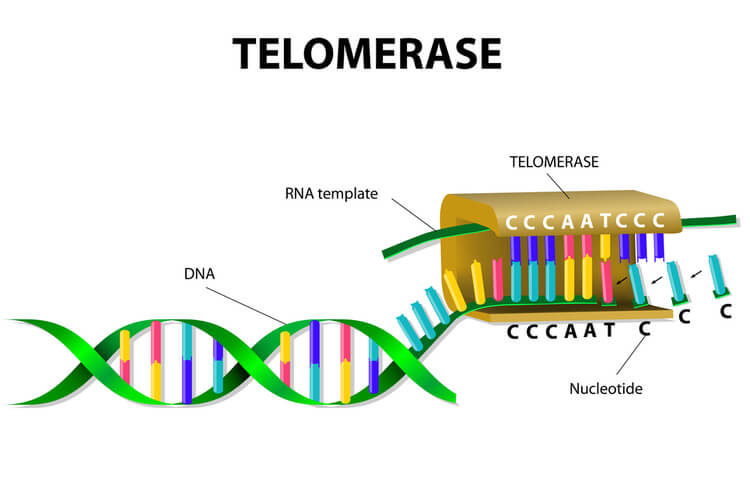
Telomere replication on the lagging strand is as follows:
- Telomerase attaches to the very end of the lagging strand, overhanging the unreplicated portion of DNA.
- Using its own RNA template, telomerase synthesizes the extending telomere, adding additional bases to the 3’ end of the lagging strand.
- Primase adds the primer on the telomere.
- DNA polymerase III binds to the primer and moves opposite of telomerase to complete the synthesis of the lagging strand.
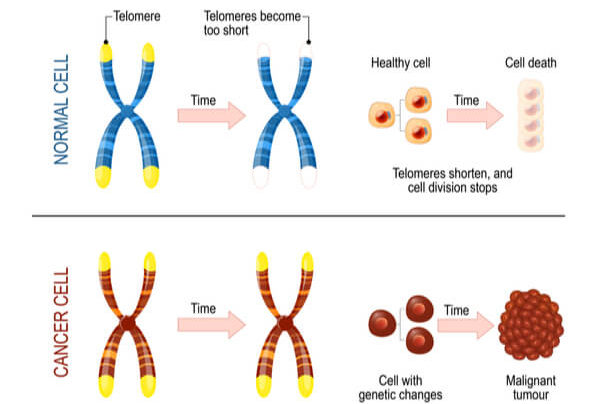
Telomeres Affect Cell Age
Telomerase is most commonly active in cell types that divide rapidly, such as with embryonic cells, stem cells, sperm cells, and immune cells. In most other cell types, telomerase activity is turned off, and telomeres become shorter with each DNA replication. This means that cells have a limited number of times that they are able to divide via mitosis before signals are sent to prevent further divisions and DNA damage. As a result, cells have age.
Research has found that increasing telomere length can also increase the lifespan of the cell. While this offers a potential treatment to growth limiting cellular diseases, it also unfortunately assists cancer persistence and survival. Approximately 90% of cancer cells have mutated to turn on telomerase activity in cell types where it should be turned off. This causes another mechanism in which cancer cells can continue to divide without control and become immortalized. Research is ongoing to determine if/how deactivating telomerase activity can either slow or stop cancer progression.
DNA Repair and Damage
Incorrect Replication
DNA replication must be fast, but it must also be extremely accurate. DNA replication occurs trillions of times in a single human. Even if there was only a single mistake in each replication, that would add up to trillions of errors that could be detrimental to the individual’s life. So how are mistakes regulated?
The first way this is done is by DNA polymerase proofreading its own work. Each complimentary pair of nucleotides has a distinct shape. Therefore, when the wrong base is placed, the shape is different enough that DNA polymerase can recognize its own mistake. DNA polymerase can then cut out this wrong match and replace it with the correct base.

While DNA polymerase is able to proofread its own work, sometimes mistakes still goes amiss. Once DNA polymerase continues down the length of the strand, mismatch repair proteins are able to edit any additional mistakes. By using markers on the old strand of DNA, the mismatch repair proteins can distinguish sequence errors on the new strand. They then remove the mismatched nucleotide and replace it accordingly. With both DNA polymerase proofreading and the mismatch repair proteins correcting additional mistakes, there is roughly only one mistake for every 1 billion nucleotides synthesized.
Environmental Damage
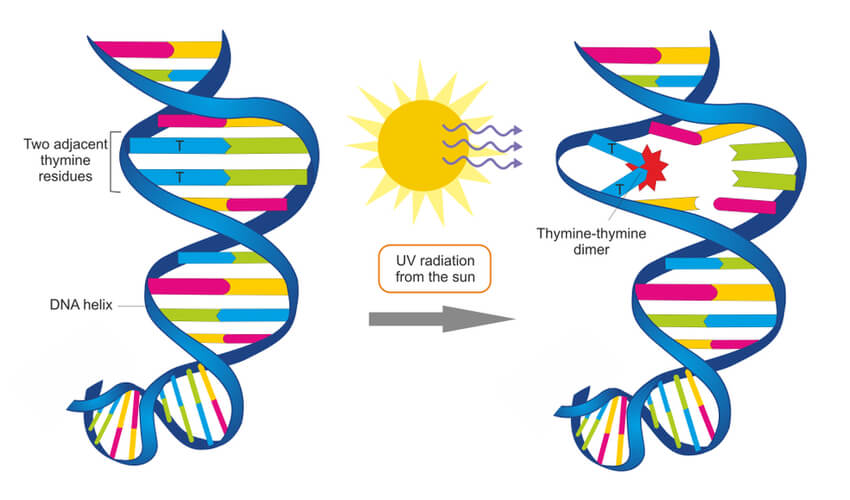
Environmental factors- such a UV radiation, X-rays, and chemical exposure- can damage DNA. For example, UV radiation found in sunlight and tanning booths can create a thymine dimer where two thymine bases next to each other form a covalent bond. This thus creates a bump in the DNA strand that prevents DNA polymerase from synthesizing past this point. These circumstances can become detrimental, and systems must be put into place to repair damages such as this.
In the case of the UV radiation, eukaryotic cells have adapted a nucleotide excision repair system that is able to detect deformities in the shape of the DNA helix. At least 18 different proteins work together to remove this deformity, using the non-damaged strand as a template to repair the damaged strand. Prokaryotic cells have a simpler but similar nucleotide excision repair system that only requires three proteins. However, for UV radiation specifically, prokaryotes use an enzyme known as photolyase to detect this damage and make repairs.
Conclusion
DNA replication is a highly regulated molecular process where a single molecule of DNA is duplicated to result in two identical DNA molecules. As a semiconservative process, the double helix is broken down into two strands, where each strand serves as the template for the newly synthesized strand by matching complementary bases. Because DNA polymerase III can only synthesize the new strands from 5′-3′, this results in a leading strand that is continuously synthesized and a lagging strand that requires the use of Okazaki fragments. Meanwhile, because eukaryotes have linear DNA, telomeres are needed to ensure genetic information is not lost during replication. Because DNA is critical to life, research continues to better understand and treat diseases caused by mutations and damages in an individual’s DNA.
Quiz
