AP Biology 5.4 - Non-Mendelian Genetics
In the previous section, we looked at how Mendel’s Laws of Inheritance allow us to predict how unlinked traits showing complete dominance are inherited. However, in most organisms, there are very few traits that show perfect complete dominance and no linkage to other traits. Since the days of Mendel, scientists have uncovered many modes of non-Mendelian genetics that change the expected genotypic and phenotypic ratios of offspring. In this section we’ll look at incomplete dominance and codominance, comparing them directly to complete dominance. Then, we’ll look at how linkage (linked genes) can break the Law of Independent Assortment. We’ll also look at how a single gene can affect multiple traits (pleiotropy) and how multiple genes can affect a single trait (polygenic trait). Finally, we’ll take a quick look at sex-linked genes, lethal alleles, and non-nuclear inheritance (mtDNA and chloroplast DNA).
Video Tutorial
The following video summarizes the most important aspects of this topic!
To watch more tutorial videos like this, please click here to see our full Youtube Channel!
Resources for this Standard
For Students & Teachers
- Overview & Video Tutorial (This article)
- Quick Test Prep
- Crossword Puzzle
For Teachers Only
ENDURING UNDERSTANDING
IST-1
Heritable information provides for the continuity of life.
LEARNING OBJECTIVE
IST-1.J
Explain deviations from Mendel’s model of the inheritance of traits.
ESSENTIAL KNOWLEDGE
IST-1.J.1
Patterns of inheritance of many traits do not follow ratios predicted by Mendel’s laws and can be identified by quantitative analysis, where observed phenotypic ratios statistically differ from the predicted ratios —
- Genes that are adjacent and close to one another on the same chromosome may appear to be genetically linked; the probability that genetically linked genes will segregate as a unit can be used to calculate the map distance between them.
IST-1.J.2
Some traits are determined by genes on sex chromosomes and are known as sex-linked traits. The pattern of inheritance of sex-linked traits can often be predicted from data, including pedigree, indicating the parent genotype/phenotype and the offspring genotypes/phenotypes.
IST-1.J.3
Many traits are the product of multiple genes and/or physiological processes acting in combination; these traits therefore do not segregate in Mendelian patterns.
IST-1.J.4
Some traits result from non-nuclear inheritance —
- Chloroplasts and mitochondria are randomly assorted to gametes and daughter cells; thus, traits determined by chloroplast and mitochondrial DNA do not follow simple Mendelian rules.
- In animals, mitochondria are transmitted by the egg and not by sperm; as such, traits determined by the mitochondrial DNA are maternally inherited.
- In plants, mitochondria and chloroplasts are transmitted in the ovule and not in the pollen; as such, mitochondria-determined and chloroplast-determined traits are maternally inherited.
5.4 Non-Mendelian Genetics Overview
Did you know that you are technically more closely related to your mom than you are to your dad? While each of your parents contributed the same number of chromosomes, all of your mitochondria and the mitochondrial DNA they carried came from your mother. Mitochondria are only inherited through egg cells since the mitochondria in sperm cells are discarded during the process of fertilization.
While normal modes of Mendelian inheritance can redistribute traits and varieties, non-Mendelian genetics can add even more variety and complexity to living organisms. Non-Mendelian genetics include different forms of dominance, like codominance and incomplete dominance, and linked genes, which are not inherited completely independently of each other. They also include genes that affect more than one trait and traits that are determined by more than one gene. Furthermore, there are also organelles like mitochondria and chloroplasts that carry their own DNA and are inherited in a much different pattern than the normal chromosomes. Elements of Non-Mendelian genetics will definitely be on the AP test. So, stick with us for a quick overview of everything you need to know about Non-Mendelian Genetics.
So, what exactly are “Non-Mendelian” genetics? This term refers to traits and genes that are not inherited in line with Mendel’s laws. Essentially, any traits that do not have alleles showing complete dominance or are inherited in unique patterns are part of non-Mendelian genetics. If you need to review standard Mendelian genetics, watch our previous video on section 5.3.
To begin, let’s take a look at two forms of non-Mendelian genetics that deal with alternative forms of dominance. Incomplete dominance happens when the heterozygote for a particular trait is a mixture of both homozygous phenotypes. For example, if a red homozygous flower and a white homozygous flower carry alleles with incomplete dominance, the hybrid resembles neither of the parents – in this case, a pink flower is produced. In fact, since neither of these alleles are dominant in the hybrid, the alleles are usually designated with a superscript – “AR” for the red allele and “AW” for the white allele.
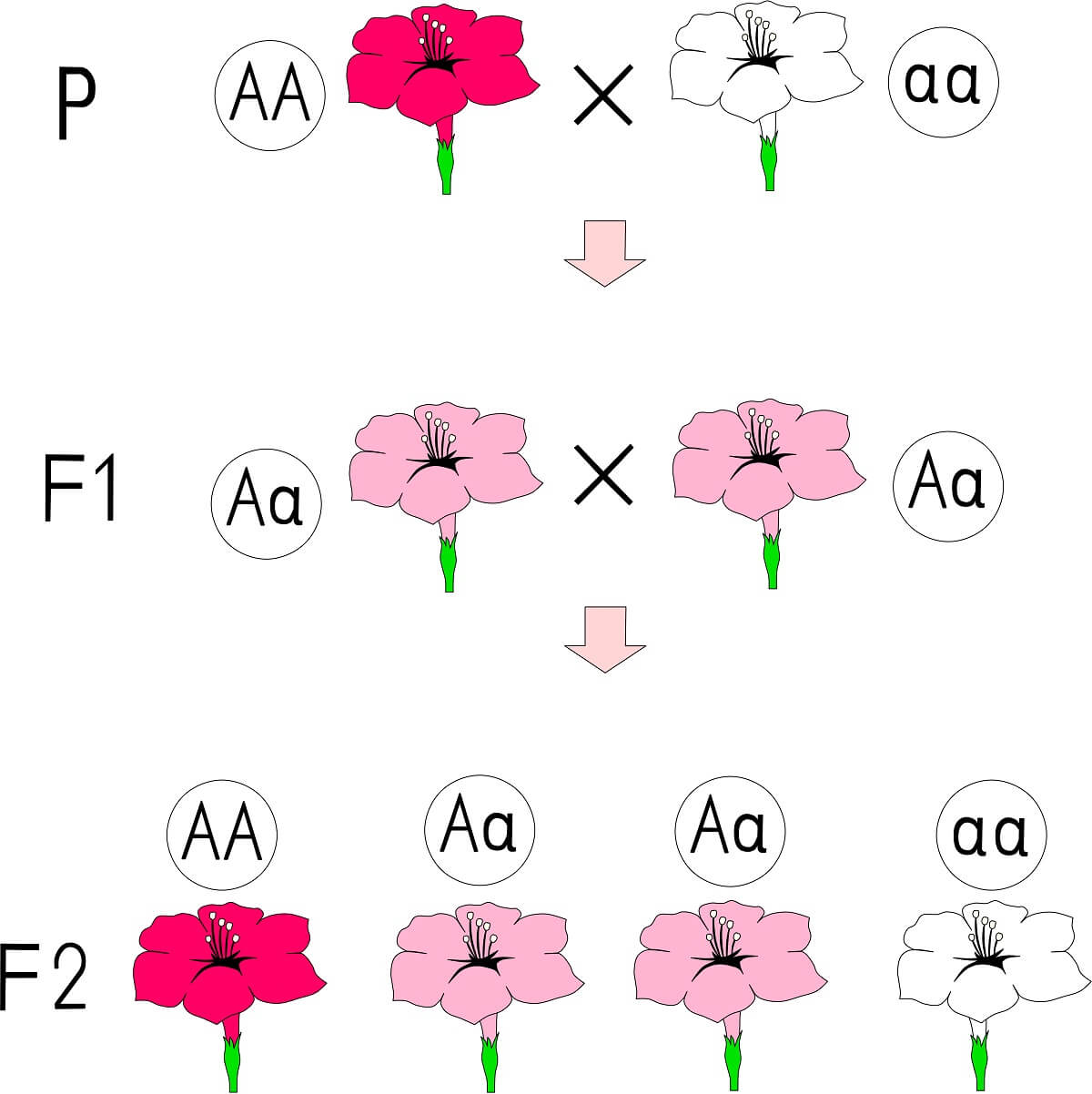
Here is what is likely happening on the molecular level. Each allele produces a different variation of the same protein. In this theoretical scenario, the red allele’s protein functions at the end of a complex pigment production pathway to convert a white pigment molecule into a red pigment molecule. The alternative white allele produces a non-functional protein, which simply passes the white pigment through. When an organism carries both of these alleles, it produces both white and red pigment molecules in equal amounts, leading to a pink flower.
In normal, Mendelian complete dominance, a very similar molecular situation could exist. However, with complete dominance, the one functional allele completely hides the recessive allele in the heterozygous phenotype. This could be because the pigment molecule created by the dominant allele is strong enough to overpower the white pigment molecule when it is present even in reduced amounts.
Now, let’s consider codominance. In codominant traits, the heterozygote is not a unique phenotype different from both homozygotes. Rather, the heterozygote shows both of the homozygous phenotypes. One classic example of codominance is the AB blood type. Blood type is also a good example of a trait with multiple alleles – A, B, and O. The A and B alleles show classic dominance over the O allele. However, when A and B are present in the same cell, both are expressed.
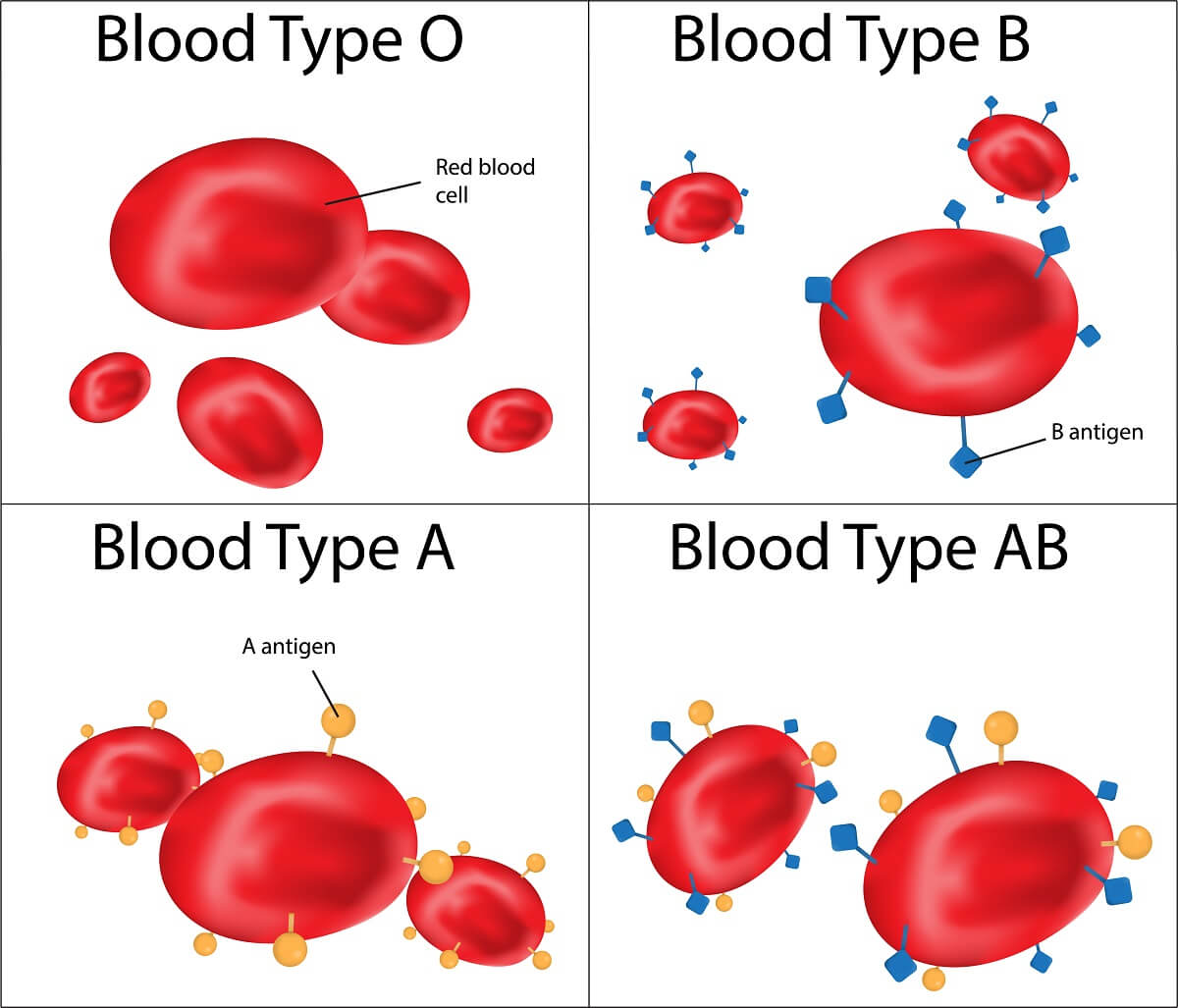
On the cellular level, this is what is happening. The O allele produces a non-functional cell-surface protein that never makes it to the cell membrane. The A and B alleles produce different functional versions of these membrane proteins. So, when both alleles are present, both proteins make it to the cell surface – making A and B codominant alleles since the heterozygote shows the phenotype of each trait separately.
You should also note that because these genes do not follow Mendel’s laws, they are not inherited in the same ratios as completely dominant traits. However, these alternate forms of dominance are not the only types of genetics that do not conform with Mendel’s laws!
Think about this… Mendel is actually quite lucky that he started his genetic research on a very purebred line of pea plants and chose the traits he did. Many other pea plant traits are governed by non-Mendelian rules, as are the traits of other plants. For example, the color of lentils is governed by multiple genes, which Mendel would have had a much harder time working out the rules for!
One of the most common violations of Mendel’s laws is linkage – when two genes are not inherited completely independently because they are located on the same chromosome. Considering that humans have over 22,000 genes and only 23 chromosomes, this is much more common than Mendel’s laws might suggest. To understand how linked genes can throw off the normal Mendelian ratios of inheritance, we have to go back to how chromosomes are separate. Keep in mind that each homologous chromosome contains the same genes, and each sister chromatid has a copy of each allele for each gene. Each of these sister chromatids will be separated into individual gametes during the process of meiosis. While genes on different chromosomes might be inherited independently, the alleles for genes on the same chromosome often end up in the same gamete.
The process of crossing over works to separate alleles on the same sister chromatid, but crossing over works best when genes are far apart on a chromosome. This process effectively mixes up alleles from homologous chromosomes, so it seems like two traits are independent of one another.
However, when genes are very close to one another, they are rarely separated by the process of crossing over. While crossing over still occurs, closely linked alleles are almost always inherited together. In fact, linked genes can help us map where different genes are located on a chromosome.
If we compare the frequency that two traits are found together in a number of offspring, we can make assumptions about how closely these two genes are located on a chromosome. For example, if we measure the frequency that eye color and blood type are inherited together, we might get something like 60%. If we measure hair color and eye color, we might get something like 10%. Since we know there is at least some linkage between these traits, we know that these traits are all on the same chromosome. But, we also know that eye color and blood type are closer together since they are inherited together more often. Essentially, genes that are closer together are separated less often by the process of crossing over. Thus, we can effectively map a chromosome, based on how far apart certain linked genes are from each other!
Now, we’re going to quickly go over a number of different types of inheritance that do not follow the typical Mendelian ratios. First, let’s consider polygenic traits – traits controlled by multiple genes. Polygenic traits are not inherited in the typical Mendelian ratios simply because there are so many different genes at play. A good example of a polygenic trait is human skin color. Skin color is controlled by several genes within the human genome and exists on a sliding scale between two extremes – similar to height, weight, and eye color.
Another non-Mendelian mode of inheritance deals with one gene controlling multiple, seemingly unrelated traits, a condition known as pleiotropy. Consider a gene in chickens that causes the feathers to be much more frizzy than normal. Peculiarly, this gene also increases the chicken’s appetite, increases the heart rate, and even delays sexual maturity. All of these things are linked to the same protein in the chicken, which is coded for by the Frizzy Gene.

Some very peculiar phenotypic ratios are obtained when lethal alleles are involved. Lethal alleles are fatal when they are inherited. Sometimes, lethal alleles are dominant (as is the case with Huntington’s disease), and sometimes they are recessive (as is the case with cystic fibrosis). When lethal alleles are fatal in embryos researchers often do not ever see these offspring. Thus, the phenotypic ratio of the offspring can be greatly affected – this gives us statistical evidence that there is likely a lethal allele involved.
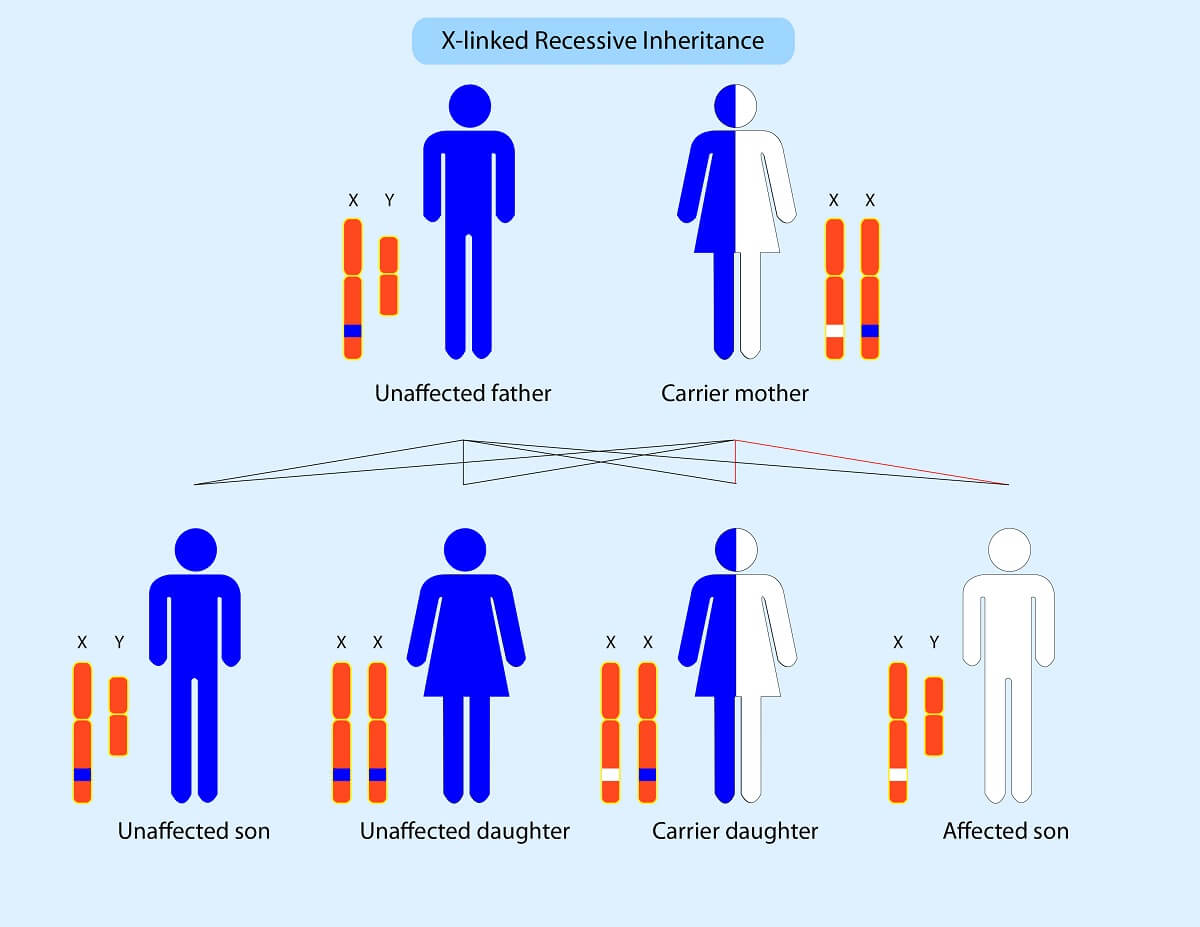
Finally, let’s consider sex-linked traits – not to be confused with linked genes. Sex-linked traits are any traits controlled by any of the genes on the sex chromosomes (X and Y in humans). Sex-linked traits alter Mendelian ratios since females receive two X chromosomes, whereas males receive one X and one Y. Unlike the other maternal and paternal chromosomes in the cell that have homologs containing the same genes, the X and Y chromosomes contain different genetic material.
For example, a sex-linked recessive disorder carried by the mother can result in carrier females, affected males, unaffected females, and unaffected males. However, if the father is the carrier, male offspring are always unaffected, since they never receive an X from the father. On the other hand, an affected father always produces carrier daughters, since they must receive the affected X chromosome. These are just some of the ways that sex-linked traits alter phenotypic ratios! Be sure to use a Punnett square if confronted with any of these unusual modes of inheritance on the test!
The final mode of non-Mendelian inheritance has to deal with the forms of DNA that are not located in the nucleus. Specifically, the DNA found within chloroplasts and mitochondria still carries important genetic information. However, these forms of DNA are not passed to the next generation in the same manner that nuclear DNA is passed. Though sperm cells do contain a number of mitochondria that provide the cell with energy, these mitochondria are not transferred to the egg cell during fertilization. This is known as non-nuclear inheritance.
The sperm cell only donates a nucleus to the new zygote, while the rest of the cell is discarded. Therefore, the only mitochondria present in a zygote are those that were already present within the egg cell. As the zygote begins dividing, these mitochondria will reproduce themselves via binary fission and be randomly assorted to each new cell.
So, unlike normal Mendelian genetics, the inheritance of mitochondrial and chloroplast DNA has no paternal component. In normal Mendelian inheritance, you receive bits of DNA from all of the humans in your line that came before you. By contrast, mitochondria only travel down the maternal line. For this reason, geneticists can trace your maternal line (also known as a maternal haplogroup) all the way back to the dawn of humanity simply by analyzing your mitochondrial DNA and the mutations it carries.
